This is a preprint.
Massively Parallel Reporter Assay Confirms Regulatory Potential of hQTLs and Reveals Important Variants in Lupus and Other Autoimmune Diseases
- PMID: 37645944
- PMCID: PMC10462090
- DOI: 10.1101/2023.08.17.553722
Massively Parallel Reporter Assay Confirms Regulatory Potential of hQTLs and Reveals Important Variants in Lupus and Other Autoimmune Diseases
Update in
-
Massively parallel reporter assay confirms regulatory potential of hQTLs and reveals important variants in lupus and other autoimmune diseases.HGG Adv. 2024 Apr 11;5(2):100279. doi: 10.1016/j.xhgg.2024.100279. Epub 2024 Feb 23. HGG Adv. 2024. PMID: 38389303 Free PMC article.
Abstract
Objective: To systematically characterize the potential for histone post-translational modifications, i.e., histone quantitative trait loci (hQTLs), expression QTLs (eQTLs), and variants on systemic lupus erythematosus (SLE) and autoimmune (AI) disease risk haplotypes to modulate gene expression in an allele dependent manner.
Methods: We designed a massively parallel reporter assay (MPRA) containing ~32K variants and transfected it into an Epstein-Barr virus transformed B cell line generated from an SLE case.
Results: Our study expands our understanding of hQTLs, illustrating that epigenetic QTLs are more likely to contribute to functional mechanisms than eQTLs and other variant types, and a large proportion of hQTLs overlap transcription start sites (TSS) of noncoding RNAs. In addition, we nominate 17 variants (including 11 novel) as putative causal variants for SLE and another 14 for various other AI diseases, prioritizing these variants for future functional studies primary and immortalized B cells.
Conclusion: We uncover important insights into the mechanistic relationships between genotype, epigenetics, gene expression, and SLE and AI disease phenotypes.
Conflict of interest statement
Conflicts of interest: None
Figures
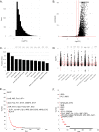
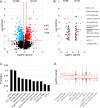
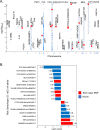


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
