Srd5a1 is Differentially Regulated and Methylated During Prepubertal Development in the Ovary and Hypothalamus
- PMID: 37646011
- PMCID: PMC10461783
- DOI: 10.1210/jendso/bvad108
Srd5a1 is Differentially Regulated and Methylated During Prepubertal Development in the Ovary and Hypothalamus
Abstract
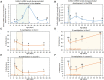
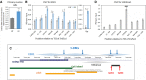
5α-reductase-1 catalyzes production of various steroids, including neurosteroids. We reported previously that expression of its encoding gene, Srd5a1, drops in murine ovaries and hypothalamic preoptic area (POA) after early-life immune stress, seemingly contributing to delayed puberty and ovarian follicle depletion, and in the ovaries the first intron was more methylated at two CpGs. Here, we hypothesized that this CpG-containing locus comprises a methylation-sensitive transcriptional enhancer for Srd5a1. We found that ovarian Srd5a1 mRNA increased 8-fold and methylation of the same two CpGs decreased up to 75% between postnatal days 10 and 30. Estradiol (E2) levels rise during this prepubertal stage, and exposure of ovarian cells to E2 increased Srd5a1 expression. Chromatin immunoprecipitation in an ovarian cell line confirmed ESR1 binding to this differentially methylated genomic region and enrichment of the enhancer modification, H3K4me1. Targeting dCas9-DNMT3 to this locus increased CpG2 methylation 2.5-fold and abolished the Srd5a1 response to E2. In the POA, Srd5a1 mRNA levels decreased 70% between postnatal days 7 and 10 and then remained constant without correlation to CpG methylation levels. Srd5a1 mRNA levels did not respond to E2 in hypothalamic GT1-7 cells, even after dCas9-TET1 reduced CpG1 methylation by 50%. The neonatal drop in POA Srd5a1 expression occurs at a time of increasing glucocorticoids, and treatment of GT1-7 cells with dexamethasone reduced Srd5a1 mRNA levels; chromatin immunoprecipitation confirmed glucocorticoid receptor binding at the enhancer. Our findings on the tissue-specific regulation of Srd5a1 and its methylation-sensitive control by E2 in the ovaries illuminate epigenetic mechanisms underlying reproductive phenotypic variation that impact life-long health.
Keywords: 5α reductase-1; Srd5a1; epigenetic; hypothalamus; methylation; ovary.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Endocrine Society.
Figures



References
-
- Bar-Sadeh B, Rudnizky S, Pnueli L, et al. . Unravelling the role of epigenetics in reproductive adaptations to early-life environment. Nat Rev Endocrinol. 2020;16(9):519‐533. - PubMed
-
- Sloboda DM, Hickey M, Hart R. Reproduction in females: the role of the early life environment. Hum Reprod Update. 2011;17(2):210‐227. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
