Structural characterization of an intrinsically disordered protein complex using integrated small-angle neutron scattering and computing
- PMID: 37646172
- PMCID: PMC10503416
- DOI: 10.1002/pro.4772
Structural characterization of an intrinsically disordered protein complex using integrated small-angle neutron scattering and computing
Abstract
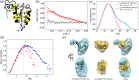
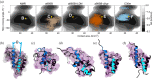
Characterizing structural ensembles of intrinsically disordered proteins (IDPs) and intrinsically disordered regions (IDRs) of proteins is essential for studying structure-function relationships. Due to the different neutron scattering lengths of hydrogen and deuterium, selective labeling and contrast matching in small-angle neutron scattering (SANS) becomes an effective tool to study dynamic structures of disordered systems. However, experimental timescales typically capture measurements averaged over multiple conformations, leaving complex SANS data for disentanglement. We hereby demonstrate an integrated method to elucidate the structural ensemble of a complex formed by two IDRs. We use data from both full contrast and contrast matching with residue-specific deuterium labeling SANS experiments, microsecond all-atom molecular dynamics (MD) simulations with four molecular mechanics force fields, and an autoencoder-based deep learning (DL) algorithm. From our combined approach, we show that selective deuteration provides additional information that helps characterize structural ensembles. We find that among the four force fields, a99SB-disp and CHARMM36m show the strongest agreement with SANS and NMR experiments. In addition, our DL algorithm not only complements conventional structural analysis methods but also successfully differentiates NMR and MD structures which are indistinguishable on the free energy surface. Lastly, we present an ensemble that describes experimental SANS and NMR data better than MD ensembles generated by one single force field and reveal three clusters of distinct conformations. Our results demonstrate a new integrated approach for characterizing structural ensembles of IDPs.
Keywords: deep learning; force field; intrinsically disordered protein; small-angle neutron scattering; structural ensemble.
© 2023 The Protein Society.
Conflict of interest statement
The authors declare no competing interests.
Figures


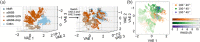

References
-
- Agarwal R, Smith MD, Smith JC. Capturing deuteration effects in a molecular mechanics force field: deuterated THF and the THF–water miscibility gap. J Chem Theory Comput. 2020;16(4):2529–2540. - PubMed
-
- Allison JR, Varnai P, Dobson CM, Vendruscolo M. Determination of the free energy landscape of α‐synuclein using spin label nuclear magnetic resonance measurements. J Am Chem Soc. 2009;131(51):18314–18326. - PubMed
-
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277(5328):965–968. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

