Machine learning-based detection of adventitious microbes in T-cell therapy cultures using long-read sequencing
- PMID: 37646508
- PMCID: PMC10580871
- DOI: 10.1128/spectrum.01350-23
Machine learning-based detection of adventitious microbes in T-cell therapy cultures using long-read sequencing
Abstract
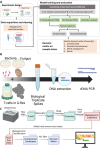
Assuring that cell therapy products are safe before releasing them for use in patients is critical. Currently, compendial sterility testing for bacteria and fungi can take 7-14 days. The goal of this work was to develop a rapid untargeted approach for the sensitive detection of microbial contaminants at low abundance from low volume samples during the manufacturing process of cell therapies. We developed a long-read sequencing methodology using Oxford Nanopore Technologies MinION platform with 16S and 18S amplicon sequencing to detect USP <71> organisms and other microbial species. Reads are classified metagenomically to predict the microbial species. We used an extreme gradient boosting machine learning algorithm (XGBoost) to first assess if a sample is contaminated, and second, determine whether the predicted contaminant is correctly classified or misclassified. The model was used to make a final decision on the sterility status of the input sample. An optimized experimental and bioinformatics pipeline starting from spiked species through to sequenced reads allowed for the detection of microbial samples at 10 colony-forming units (CFU)/mL using metagenomic classification. Machine learning can be coupled with long-read sequencing to detect and identify sample sterility status and microbial species present in T-cell cultures, including the USP <71> organisms to 10 CFU/mL. IMPORTANCE This research presents a novel method for rapidly and accurately detecting microbial contaminants in cell therapy products, which is essential for ensuring patient safety. Traditional testing methods are time-consuming, taking 7-14 days, while our approach can significantly reduce this time. By combining advanced long-read nanopore sequencing techniques and machine learning, we can effectively identify the presence and types of microbial contaminants at low abundance levels. This breakthrough has the potential to improve the safety and efficiency of cell therapy manufacturing, leading to better patient outcomes and a more streamlined production process.
Keywords: T-cells; adventitious agents; machine learning; sterility.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- García-Bernal D, García-Arranz M, Yáñez RM, Hervás-Salcedo R, Cortés A, Fernández-García M, Hernando-Rodríguez M, Quintana-Bustamante Ó, Bueren JA, García-Olmo D, Moraleda JM, Segovia JC, Zapata AG. 2021. The current status of mesenchymal stromal cells: controversies, unresolved issues and some promising solutions to improve their therapeutic efficacy. Front Cell Dev Biol 9:650664. doi: 10.3389/fcell.2021.650664 - DOI - PMC - PubMed
-
- Barone PW, Wiebe ME, Leung JC, Hussein ITM, Keumurian FJ, Bouressa J, Brussel A, Chen D, Chong M, Dehghani H, Gerentes L, Gilbert J, Gold D, Kiss R, Kreil TR, Labatut R, Li Y, Müllberg J, Mallet L, Menzel C, Moody M, Monpoeho S, Murphy M, Plavsic M, Roth NJ, Roush D, Ruffing M, Schicho R, Snyder R, Stark D, Zhang C, Wolfrum J, Sinskey AJ, Springs SL. 2020. Viral contamination in biologic manufacture and implications for emerging therapies. Nat Biotechnol 38:563–572. doi: 10.1038/s41587-020-0507-2 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

