A novel ruthenium-silver based antimicrobial potentiates aminoglycoside activity against Pseudomonas aeruginosa
- PMID: 37646510
- PMCID: PMC10597350
- DOI: 10.1128/msphere.00190-23
A novel ruthenium-silver based antimicrobial potentiates aminoglycoside activity against Pseudomonas aeruginosa
Abstract
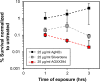
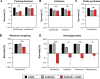
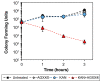
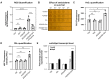
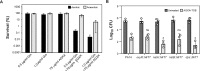
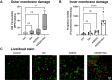
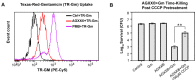
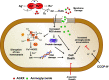
The rapid dissemination of antibiotic resistance combined with the decline in the discovery of novel antibiotics represents a major challenge for infectious disease control that can only be mitigated by investments in novel treatment strategies. Alternative antimicrobials, including silver, have regained interest due to their diverse mechanisms of inhibiting microbial growth. One such example is AGXX, a broad-spectrum antimicrobial that produces highly cytotoxic reactive oxygen species (ROS) to inflict extensive macromolecular damage. Due to the connections identified between ROS production and antibiotic lethality, we hypothesized that AGXX could potentially increase the activity of conventional antibiotics. Using the gram-negative pathogen Pseudomonas aeruginosa, we screened possible synergistic effects of AGXX on several antibiotic classes. We found that the combination of AGXX and aminoglycosides tested at sublethal concentrations led to a rapid exponential decrease in bacterial survival and restored the sensitivity of a kanamycin-resistant strain. ROS production contributes significantly to the bactericidal effects of AGXX/aminoglycoside treatments, which is dependent on oxygen availability and can be reduced by the addition of ROS scavengers. Additionally, P. aeruginosa strains deficient in ROS detoxifying/repair genes were more susceptible to AGXX/aminoglycoside treatment. We further demonstrate that this synergistic interaction was associated with a significant increase in outer and inner membrane permeability, resulting in increased antibiotic influx. Our study also revealed that AGXX/aminoglycoside-mediated killing requires an active proton motive force across the bacterial membrane. Overall, our findings provide an understanding of cellular targets that could be inhibited to increase the activity of conventional antimicrobials. IMPORTANCE The emergence of drug-resistant bacteria coupled with the decline in antibiotic development highlights the need for novel alternatives. Thus, new strategies aimed at repurposing conventional antibiotics have gained significant interest. The necessity of these interventions is evident especially in gram-negative pathogens as they are particularly difficult to treat due to their outer membrane. This study highlights the effectiveness of the antimicrobial AGXX in potentiating aminoglycoside activities against P. aeruginosa. The combination of AGXX and aminoglycosides not only reduces bacterial survival rapidly but also significantly re-sensitizes aminoglycoside-resistant P. aeruginosa strains. In combination with gentamicin, AGXX induces increased endogenous oxidative stress, membrane damage, and iron-sulfur cluster disruption. These findings emphasize AGXX's potential as a route of antibiotic adjuvant development and shed light on potential targets to enhance aminoglycoside activity.
Keywords: aminoglycosides; antibiotics; iron; membranes; reactive oxygen species; redox stress; silver.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Update of
-
The Novel Silver-Containing Antimicrobial Potentiates Aminoglycoside Activity Against Pseudomonas aeruginosa.bioRxiv [Preprint]. 2023 Jun 29:2023.03.15.532855. doi: 10.1101/2023.03.15.532855. bioRxiv. 2023. Update in: mSphere. 2023 Oct 24;8(5):e0019023. doi: 10.1128/msphere.00190-23. PMID: 36993297 Free PMC article. Updated. Preprint.
Similar articles
-
A novel silver-ruthenium-based antimicrobial kills Gram-negative bacteria through oxidative stress-induced macromolecular damage.mSphere. 2025 Jun 25;10(6):e0001725. doi: 10.1128/msphere.00017-25. Epub 2025 May 30. mSphere. 2025. PMID: 40444966 Free PMC article.
-
The Novel Silver-Containing Antimicrobial Potentiates Aminoglycoside Activity Against Pseudomonas aeruginosa.bioRxiv [Preprint]. 2023 Jun 29:2023.03.15.532855. doi: 10.1101/2023.03.15.532855. bioRxiv. 2023. Update in: mSphere. 2023 Oct 24;8(5):e0019023. doi: 10.1128/msphere.00190-23. PMID: 36993297 Free PMC article. Updated. Preprint.
-
Rapid Freezing Enables Aminoglycosides To Eradicate Bacterial Persisters via Enhancing Mechanosensitive Channel MscL-Mediated Antibiotic Uptake.mBio. 2020 Feb 11;11(1):e03239-19. doi: 10.1128/mBio.03239-19. mBio. 2020. PMID: 32047133 Free PMC article.
-
Unlocking Enhanced Efficacy of Aminoglycoside Antibiotics Against Pseudomonas aeruginosa.Microb Biotechnol. 2025 Jun;18(6):e70174. doi: 10.1111/1751-7915.70174. Microb Biotechnol. 2025. PMID: 40448301 Free PMC article. Review.
-
Aminoglycosides plus beta-lactams against gram-negative organisms. Evaluation of in vitro synergy and chemical interactions.Am J Med. 1986 Jun 30;80(6B):126-37. doi: 10.1016/0002-9343(86)90490-0. Am J Med. 1986. PMID: 3088998 Review.
Cited by
-
Identification and characterization of a novel chromosome-encoded aminoglycoside O-nucleotidyltransferase gene, ant(9)-Id, in Providencia sp. TYF-12 isolated from the marine fish intestine.Front Microbiol. 2024 Dec 12;15:1475172. doi: 10.3389/fmicb.2024.1475172. eCollection 2024. Front Microbiol. 2024. PMID: 39726966 Free PMC article.
-
Aminoglycoside uptake, stress, and potentiation in Gram-negative bacteria: new therapies with old molecules.Microbiol Mol Biol Rev. 2023 Dec 20;87(4):e0003622. doi: 10.1128/mmbr.00036-22. Epub 2023 Dec 4. Microbiol Mol Biol Rev. 2023. PMID: 38047635 Free PMC article. Review.
-
A Novel Silver-Ruthenium-Based Antimicrobial Kills Gram-Negative Bacteria Through Oxidative Stress-Induced Macromolecular Damage.bioRxiv [Preprint]. 2025 Jan 4:2025.01.03.631245. doi: 10.1101/2025.01.03.631245. bioRxiv. 2025. Update in: mSphere. 2025 Jun 25;10(6):e0001725. doi: 10.1128/msphere.00017-25. PMID: 39803548 Free PMC article. Updated. Preprint.
-
A novel silver-ruthenium-based antimicrobial kills Gram-negative bacteria through oxidative stress-induced macromolecular damage.mSphere. 2025 Jun 25;10(6):e0001725. doi: 10.1128/msphere.00017-25. Epub 2025 May 30. mSphere. 2025. PMID: 40444966 Free PMC article.
-
Exogenous NADH promotes the bactericidal effect of aminoglycoside antibiotics against Edwardsiella tarda.Virulence. 2024 Dec;15(1):2367647. doi: 10.1080/21505594.2024.2367647. Epub 2024 Jun 17. Virulence. 2024. PMID: 38884466 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

