Analytic validation of NeXT Dx™, a comprehensive genomic profiling assay
- PMID: 37646774
- PMCID: PMC10467627
- DOI: 10.18632/oncotarget.28490
Analytic validation of NeXT Dx™, a comprehensive genomic profiling assay
Abstract
We describe the analytic validation of NeXT Dx, a comprehensive genomic profiling assay to aid therapy and clinical trial selection for patients diagnosed with solid tumor cancers. Proprietary methods were utilized to perform whole exome and whole transcriptome sequencing for detection of single nucleotide variants (SNVs), insertions/deletions (indels), copy number alterations (CNAs), and gene fusions, and determination of tumor mutation burden and microsatellite instability. Variant calling is enhanced by sequencing a patient-specific normal sample from, for example, a blood specimen. This provides highly accurate somatic variant calls as well as the incidental reporting of pathogenic and likely pathogenic germline alterations. Fusion detection via RNA sequencing provides more extensive and accurate fusion calling compared to DNA-based tests. NeXT Dx features the proprietary Accuracy and Content Enhanced technology, developed to optimize sequencing and provide more uniform coverage across the exome. The exome was validated at a median sequencing depth of >500x. While variants from 401 cancer-associated genes are currently reported from the assay, the exome/transcriptome assay is broadly validated to enable reporting of additional variants as they become clinically relevant. NeXT Dx demonstrated analytic sensitivities as follows: SNVs (99.4%), indels (98.2%), CNAs (98.0%), and fusions (95.8%). The overall analytic specificity was >99.0%.
Keywords: comprehensive genomic profiling; precision medicine; tumor-normal; whole exome sequencing; whole transcriptome sequencing.
Conflict of interest statement
All authors have or had a financial relationship as employees of Personalis, Inc.
Figures
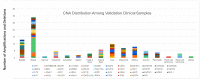

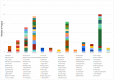

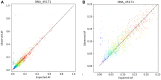

References
-
- American Cancer Society. https://www.cancer.org/. Accessed Jan. 2023.
-
- El-Deiry WS, Goldberg RM, Lenz HJ, Shields AF, Gibney GT, Tan AR, Brown J, Eisenberg B, Heath EI, Phuphanich S, Kim E, Brenner AJ, Marshall JL. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J Clin. 2019; 69:305–43. 10.3322/caac.21560. - DOI - PMC - PubMed
-
- Wheler JJ, Janku F, Naing A, Li Y, Stephen B, Zinner R, Subbiah V, Fu S, Karp D, Falchook GS, Tsimberidou AM, Piha-Paul S, Anderson R, et al. Cancer Therapy Directed by Comprehensive Genomic Profiling: A Single Center Study. Cancer Res. 2016; 76:3690–701. 10.1158/0008-5472.CAN-15-3043. - DOI - PubMed
-
- Gutierrez ME, Choi K, Lanman RB, Licitra EJ, Skrzypczak SM, Pe Benito R, Wu T, Arunajadai S, Kaur S, Harper H, Pecora AL, Schultz EV, Goldberg SL. Genomic Profiling of Advanced Non-Small Cell Lung Cancer in Community Settings: Gaps and Opportunities. Clin Lung Cancer. 2017; 18:651–59. 10.1016/j.cllc.2017.04.004. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

