The molecular epidemiology and antimicrobial resistance of Staphylococcus pseudintermedius canine clinical isolates submitted to a veterinary diagnostic laboratory in South Africa
- PMID: 37647319
- PMCID: PMC10468042
- DOI: 10.1371/journal.pone.0290645
The molecular epidemiology and antimicrobial resistance of Staphylococcus pseudintermedius canine clinical isolates submitted to a veterinary diagnostic laboratory in South Africa
Abstract
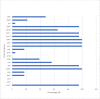
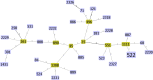
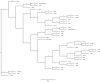
Staphylococcus pseudintermedius is an important cause of clinical infections in small-animal-veterinary medicine. Evolutionary changes of strains using multilocus sequence typing (MLST) have been observed among S. pseudintermedius in European countries and the United States. However, there are limited or no studies on the detection of methicillin resistant Staphylococcus pseudintermedius (MRSP) and predominating MLST strains in South Africa. Therefore, this study aimed to determine the molecular epidemiology of S. pseudintermedius in South Africa. Twenty-six, non-duplicate, clinical isolates from dogs were obtained as convenience samples from four provinces in South Africa. The Kirby Bauer disk diffusion test was used to determine antimicrobial susceptibility. We used Resfinder and the Comprehensive Antibiotic Resistance Database (CARD) to detect antimicrobial resistance genes. Virulence genes were identified using the virulence factor database and Basic Local Alignment Search Tool (BLASTN) on Geneious prime. geoBURST analysis was used to study relationships between MLST. Finally, the maximum likelihood phylogeny was determined using Randomized Axelerated Maximum Likelihood (RAxML). Twenty-three isolates were confirmed as S. pseudintermedius of which 14 were MRSP. In addition to β-lactam antimicrobials, MRSP isolates were resistant to tetracycline (85.7%), doxycycline (92.8%), kanamycin (92.8%), and gentamicin (85.7%). The isolates harbored antimicrobial resistance genes (tetM, ermB, drfG, cat, aac(6')-Ie-aph(2")-Ia, ant(6)-Ia, and aph(3')-III) and virulence genes (AdsA, geh, icaA, and lip). MLST analysis showed that ST2228, ST2229, ST2230, ST2231, ST2232, ST2318, ST2326 and ST2327 are unique sequence types in South Africa. Whereas, previously reported major STs including ST45, ST71, ST181, ST551 and ST496 were also detected. The geoBURST and phylogenetic analysis suggests that the isolates in South Africa are likely genetically related to isolates identified in other countries. Highly resistant MRSP strains (ST496, ST71, and ST45) were reported that could present challenges in the treatment of canine infections in South Africa. Hence, we have gained a better understanding of the epidemiology of MRSP in the African continent, the genes involved in resistance and virulence factors associated with these organisms.
Copyright: © 2023 Phophi et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Temporal changes in antibiotic resistance and population structure of methicillin-resistant Staphylococcus pseudintermedius between 2010 and 2021 in the United States.Comp Immunol Microbiol Infect Dis. 2023 Sep;100:102028. doi: 10.1016/j.cimid.2023.102028. Epub 2023 Jul 24. Comp Immunol Microbiol Infect Dis. 2023. PMID: 37517211
-
Clonal diversity and geographic distribution of methicillin-resistant Staphylococcus pseudintermedius from Australian animals: Discovery of novel sequence types.Vet Microbiol. 2018 Jan;213:58-65. doi: 10.1016/j.vetmic.2017.11.018. Epub 2017 Nov 21. Vet Microbiol. 2018. PMID: 29292005
-
Identification and molecular epidemiology of methicillin resistant Staphylococcus pseudintermedius strains isolated from canine clinical samples in Argentina.BMC Vet Res. 2019 Jul 27;15(1):264. doi: 10.1186/s12917-019-1990-x. BMC Vet Res. 2019. PMID: 31351494 Free PMC article.
-
Methicillin-resistant Staphylococcus pseudintermedius: epidemiological changes, antibiotic resistance, and alternative therapeutic strategies.Vet Res Commun. 2024 Dec;48(6):3505-3515. doi: 10.1007/s11259-024-10508-8. Epub 2024 Aug 21. Vet Res Commun. 2024. PMID: 39167258 Free PMC article. Review.
-
Systematic Review on Global Epidemiology of Methicillin-Resistant Staphylococcus pseudintermedius: Inference of Population Structure from Multilocus Sequence Typing Data.Front Microbiol. 2016 Oct 18;7:1599. doi: 10.3389/fmicb.2016.01599. eCollection 2016. Front Microbiol. 2016. PMID: 27803691 Free PMC article. Review.
Cited by
-
Whole Genome Sequencing and Comparative Genomics of Six Staphylococcus schleiferi and Staphylococcus coagulans Isolates.Genes (Basel). 2024 Feb 24;15(3):284. doi: 10.3390/genes15030284. Genes (Basel). 2024. PMID: 38540343 Free PMC article.
-
Genotypic profile of Staphylococcus spp., Enterococcus spp., and E. coli colonizing dogs, surgeons, and environment during the intraoperative period: a cross-sectional study in a veterinary teaching hospital in Brazil.BMC Vet Res. 2025 Mar 6;21(1):147. doi: 10.1186/s12917-025-04611-4. BMC Vet Res. 2025. PMID: 40050816 Free PMC article.
-
Companions in antimicrobial resistance: examining transmission of common antimicrobial-resistant organisms between people and their dogs, cats, and horses.Clin Microbiol Rev. 2025 Mar 13;38(1):e0014622. doi: 10.1128/cmr.00146-22. Epub 2025 Jan 24. Clin Microbiol Rev. 2025. PMID: 39853095 Review.
-
Emergence of novel methicillin resistant Staphylococcus pseudintermedius lineages revealed by whole genome sequencing of isolates from companion animals and humans in Scotland.PLoS One. 2024 Jul 5;19(7):e0305211. doi: 10.1371/journal.pone.0305211. eCollection 2024. PLoS One. 2024. PMID: 38968222 Free PMC article.
-
Genome Mining Reveals a Sactipeptide Biosynthetic Cluster in Staphylococcus pseudintermedius.Vet Sci. 2025 Jul 2;12(7):635. doi: 10.3390/vetsci12070635. Vet Sci. 2025. PMID: 40711295 Free PMC article.
References
-
- Videla R, Solyman SM, Brahmbhatt A, Sadeghi L, Bemis DA, Kania SA. Clonal Complexes and Antimicrobial Susceptibility Profiles of Staphylococcus pseudintermedius Isolates from Dogs in the United States. Microb Drug Resist Larchmt N. 2018. Jan 1;24(1):83–8. - PubMed
-
- Somayaji R, Priyantha MAR, Rubin JE, Church D. Human infections due to Staphylococcus pseudintermedius, an emerging zoonosis of canine origin: report of 24 cases. Diagn Microbiol Infect Dis. 2016. Aug 1;85(4):471–6. - PubMed
-
- Marsilio F, Di Francesco CE, Di Martino B. Coagulase-Positive and Coagulase-Negative Staphylococci Animal Diseases. In: Pet-to-Man Travelling Staphylococci: A World in Progress. Elsevier Inc.; 2018. p. 43–50.
-
- Dos Santos TP, Damborg P, Moodley A, Guardabassi L. Systematic review on global epidemiology of methicillin-resistant Staphylococcus pseudintermedius: Inference of population structure from multilocus sequence typing data. Vol. 7, Frontiers in Microbiology. Frontiers Media S.A.; 2016. - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

