A blood-based marker of mitochondrial DNA damage in Parkinson's disease
- PMID: 37647388
- PMCID: PMC11135133
- DOI: 10.1126/scitranslmed.abo1557
A blood-based marker of mitochondrial DNA damage in Parkinson's disease
Abstract
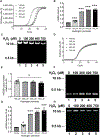
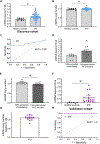
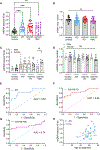
Parkinson's disease (PD) is the most common neurodegenerative movement disorder, and neuroprotective or disease-modifying interventions remain elusive. High-throughput markers aimed at stratifying patients on the basis of shared etiology are required to ensure the success of disease-modifying therapies in clinical trials. Mitochondrial dysfunction plays a prominent role in the pathogenesis of PD. Previously, we found brain region-specific accumulation of mitochondrial DNA (mtDNA) damage in PD neuronal culture and animal models, as well as in human PD postmortem brain tissue. To investigate mtDNA damage as a potential blood-based marker for PD, we describe herein a PCR-based assay (Mito DNADX) that allows for the accurate real-time quantification of mtDNA damage in a scalable platform. We found that mtDNA damage was increased in peripheral blood mononuclear cells derived from patients with idiopathic PD and those harboring the PD-associated leucine-rich repeat kinase 2 (LRRK2) G2019S mutation in comparison with age-matched controls. In addition, mtDNA damage was elevated in non-disease-manifesting LRRK2 mutation carriers, demonstrating that mtDNA damage can occur irrespective of a PD diagnosis. We further established that Lrrk2 G2019S knock-in mice displayed increased mtDNA damage, whereas Lrrk2 knockout mice showed fewer mtDNA lesions in the ventral midbrain, compared with wild-type control mice. Furthermore, a small-molecule kinase inhibitor of LRRK2 mitigated mtDNA damage in a rotenone PD rat midbrain neuron model and in idiopathic PD patient-derived lymphoblastoid cell lines. Quantifying mtDNA damage using the Mito DNADX assay may have utility as a candidate marker of PD and for measuring the pharmacodynamic response to LRRK2 kinase inhibitors.
Conflict of interest statement
Figures



References
-
- Fearnley JM, Lees AJ, Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain 114 (Pt 5), 2283–2301 (1991). - PubMed
-
- Marras C, Lang A, Parkinson’s disease subtypes: Lost in translation? J. Neurol. Neurosurg. Psychiatry 84, 409–415 (2013). - PubMed
-
- Mestre TA, Fereshtehnejad SM, Berg D, Bohnen NI, Dujardin K, Erro R, Espay AJ, Halliday G, van Hilten JJ, Hu MT, Jeon B, Klein C, Leentjens AFG, Marinus J, Mollenhauer B, Postuma R, Rajalingam R, Rodriguez-Violante M, Simuni T, Surmeier DJ, Weintraub D, McDermott MP, Lawton M, Marras C, Parkinson’s disease subtypes: Critical appraisal and recommendations. J. Parkinsons Dis. 11, 395–404 (2021). - PMC - PubMed
-
- Espay AJ, Schwarzschild MA, Tanner CM, Fernandez HH, Simon DK, Leverenz JB, Merola A, Chen-Plotkin A, Brundin P, Kauffman MA, Erro R, Kieburtz K, Woo D, Macklin EA, Standaert DG, Lang AE, Biomarker-driven phenotyping in Parkinson’s disease: A translational missing link in disease-modifying clinical trials. Mov. Disord. 32, 319–324 (2017). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

