Leaf metabolic traits reveal hidden dimensions of plant form and function
- PMID: 37647404
- PMCID: PMC10468135
- DOI: 10.1126/sciadv.adi4029
Leaf metabolic traits reveal hidden dimensions of plant form and function
Abstract
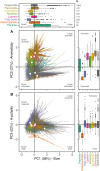
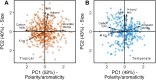
The metabolome is the biochemical basis of plant form and function, but we know little about its macroecological variation across the plant kingdom. Here, we used the plant functional trait concept to interpret leaf metabolome variation among 457 tropical and 339 temperate plant species. Distilling metabolite chemistry into five metabolic functional traits reveals that plants vary on two major axes of leaf metabolic specialization-a leaf chemical defense spectrum and an expression of leaf longevity. Axes are similar for tropical and temperate species, with many trait combinations being viable. However, metabolic traits vary orthogonally to life-history strategies described by widely used functional traits. The metabolome thus expands the functional trait concept by providing additional axes of metabolic specialization for examining plant form and function.
Figures


References
-
- Wang S., Alseekh S., Fernie A. R., Luo J., The structure and function of major plant metabolite modifications. Mol. Plant 12, 899–919 (2019). - PubMed
-
- Klee H. J., Tieman D. M., The genetics of fruit flavour preferences. Nat. Rev. Genet. 19, 347–356 (2018). - PubMed
-
- Schmidt B. M., Ribnicky D. M., Lipsky P. E., Raskin I., Revisiting the ancient concept of botanical therapeutics. Nat. Chem. Biol. 3, 360–366 (2007). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

