HAPLN1 matrikine: a bone marrow homing factor linked to poor outcomes in patients with MM
- PMID: 37647592
- PMCID: PMC10685165
- DOI: 10.1182/bloodadvances.2023010139
HAPLN1 matrikine: a bone marrow homing factor linked to poor outcomes in patients with MM
Abstract
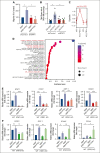
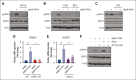
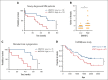
The bone marrow (BM) microenvironment is critical for dissemination, growth, and survival of multiple myeloma (MM) cells. Homing of myeloma cells to the BM niche is a crucial step in MM dissemination, but the mechanisms involved are incompletely understood. In particular, any role of matrikines, neofunctional peptides derived from extracellular matrix proteins, remains unknown. Here, we report that a matrikine derived from hyaluronan and proteoglycan link protein 1 (HAPLN1) induces MM cell adhesion to the BM stromal components, such as fibronectin, endothelial cells, and stromal cells and, furthermore, induces their chemotactic and chemokinetic migration. In a mouse xenograft model, we show that MM cells preferentially home to HAPLN1 matrikine-conditioned BM. The transcription factor STAT1 is activated by HAPLN1 matrikine and is necessary to induce MM cell adhesion, migration, migration-related genes, and BM homing. STAT1 activation is mediated by interferon beta (IFN-β), which is induced by NF-κB after stimulation by HAPLN1 matrikine. Finally, we also provide evidence that higher levels of HAPLN1 in BM samples correlate with poorer progression-free survival of patients with newly diagnosed MM. These data reveal that a matrikine present in the BM microenvironment acts as a chemoattractant, plays an important role in BM homing of MM cells via NF-κB-IFN-β-STAT1 signaling, and may help identify patients with poor outcomes. This study also provides a mechanistic rationale for targeting HAPLN1 matrikine in MM therapy.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures
Similar articles
-
A Hyaluronan and Proteoglycan Link Protein 1 Matrikine: Role of Matrix Metalloproteinase 2 in Multiple Myeloma NF-κB Activation and Drug Resistance.Mol Cancer Res. 2022 Sep 2;20(9):1456-1466. doi: 10.1158/1541-7786.MCR-21-0941. Mol Cancer Res. 2022. PMID: 35604822 Free PMC article.
-
Hyaluronan and proteoglycan link protein 1 (HAPLN1) activates bortezomib-resistant NF-κB activity and increases drug resistance in multiple myeloma.J Biol Chem. 2018 Feb 16;293(7):2452-2465. doi: 10.1074/jbc.RA117.000667. Epub 2017 Dec 26. J Biol Chem. 2018. PMID: 29279332 Free PMC article.
-
Ectopic CH60 mediates HAPLN1-induced cell survival signaling in multiple myeloma.Life Sci Alliance. 2022 Dec 13;6(3):e202201636. doi: 10.26508/lsa.202201636. Print 2023 Mar. Life Sci Alliance. 2022. PMID: 36625202 Free PMC article.
-
Migration, adhesion and differentiation of malignant plasma cells in the 5T murine model of myeloma.Verh K Acad Geneeskd Belg. 2003;65(2):127-34. Verh K Acad Geneeskd Belg. 2003. PMID: 12870183 Review.
-
Role of NF-κB Signaling in the Interplay between Multiple Myeloma and Mesenchymal Stromal Cells.Int J Mol Sci. 2023 Jan 17;24(3):1823. doi: 10.3390/ijms24031823. Int J Mol Sci. 2023. PMID: 36768145 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

