CRISPR screens decode cancer cell pathways that trigger γδ T cell detection
- PMID: 37648854
- PMCID: PMC11003766
- DOI: 10.1038/s41586-023-06482-x
CRISPR screens decode cancer cell pathways that trigger γδ T cell detection
Abstract
γδ T cells are potent anticancer effectors with the potential to target tumours broadly, independent of patient-specific neoantigens or human leukocyte antigen background1-5. γδ T cells can sense conserved cell stress signals prevalent in transformed cells2,3, although the mechanisms behind the targeting of stressed target cells remain poorly characterized. Vγ9Vδ2 T cells-the most abundant subset of human γδ T cells4-recognize a protein complex containing butyrophilin 2A1 (BTN2A1) and BTN3A1 (refs. 6-8), a widely expressed cell surface protein that is activated by phosphoantigens abundantly produced by tumour cells. Here we combined genome-wide CRISPR screens in target cancer cells to identify pathways that regulate γδ T cell killing and BTN3A cell surface expression. The screens showed previously unappreciated multilayered regulation of BTN3A abundance on the cell surface and triggering of γδ T cells through transcription, post-translational modifications and membrane trafficking. In addition, diverse genetic perturbations and inhibitors disrupting metabolic pathways in the cancer cells, particularly ATP-producing processes, were found to alter BTN3A levels. This induction of both BTN3A and BTN2A1 during metabolic crises is dependent on AMP-activated protein kinase (AMPK). Finally, small-molecule activation of AMPK in a cell line model and in patient-derived tumour organoids led to increased expression of the BTN2A1-BTN3A complex and increased Vγ9Vδ2 T cell receptor-mediated killing. This AMPK-dependent mechanism of metabolic stress-induced ligand upregulation deepens our understanding of γδ T cell stress surveillance and suggests new avenues available to enhance γδ T cell anticancer activity.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
A.M. is a co-founder of Arsenal Biosciences, Spotlight Therapeutics, and Survey Genomics, serves on the boards of directors at Spotlight Therapeutics and Survey Genomics, is a board observer (and former member of the board of directors) at Arsenal Biosciences, is a member of the scientific advisory boards of Arsenal Biosciences, Spotlight Therapeutics, Survey Genomics, NewLimit, Amgen, Lightcast, and Tenaya, owns stock in Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics, PACT Pharma, Lightcast, and Tenaya, and has received fees from Arsenal Biosciences, Spotlight Therapeutics, NewLimit, Survey Genomics, Tenaya, Lightcast, 23andMe, PACT Pharma, Juno Therapeutics, Trizell, Vertex, Merck, Amgen, Genentech, AlphaSights, Rupert Case Management, Bernstein, and ALDA. A.M. is an investor in and informal advisor to Offline Ventures and a client of EPIQ. J.W.F. was a consultant for NewLimit, is an employee of Genentech, and has equity in Roche. The Marson laboratory has received research support from Juno Therapeutics, Epinomics, Sanofi, GlaxoSmithKline, Gilead, and Anthem. J.K. is a shareholder of Gadeta B.V.. J.K. and Z.S. are inventors on patents with γδTCR related topics. A.M. and M.R.M. are inventors on patent applications that have been filed based on the findings described here.
Figures





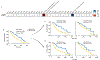






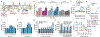


Comment in
-
Unlocking γδ T cell power: pathways that boost cancer defense.Mol Biomed. 2024 Feb 5;5(1):5. doi: 10.1186/s43556-023-00168-6. Mol Biomed. 2024. PMID: 38311633 Free PMC article. No abstract available.
References
-
- Silva-Santos B, Serre K & Norell H γδ T cells in cancer. Nat. Rev. Immunol 15, 683–691 (2015). - PubMed
-
- Sebestyen Z, Prinz I, Déchanet-Merville J, Silva-Santos B & Kuball J Translating gammadelta (γδ) T cells and their receptors into cancer cell therapies. Nat. Rev. Drug Discov 19, 169–184 (2020). - PubMed
-
- Mensurado S, Blanco-Domínguez R & Silva-Santos B The emerging roles of γδ T cells in cancer immunotherapy. Nat. Rev. Clin. Oncol 20, 178–191 (2023). - PubMed
Methods References
-
- Jiang S et al. CRISPR/Cas9-Mediated Genome Editing in Epstein-Barr Virus-Transformed Lymphoblastoid B-Cell Lines. Curr. Protoc. Mol. Biol 121, 31.12.1–31.12.23 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

