Metagenomic and culture-dependent approaches unveil active microbial community and novel functional genes involved in arsenic mobilization and detoxification in groundwater
- PMID: 37648982
- PMCID: PMC10466822
- DOI: 10.1186/s12866-023-02980-0
Metagenomic and culture-dependent approaches unveil active microbial community and novel functional genes involved in arsenic mobilization and detoxification in groundwater
Abstract
Background: Arsenic (As) and its species are major pollutants in ecological bodied including groundwater in Bangladesh rendering serious public health concern. Bacteria with arsenotrophic genes have been found in the aquifer, converting toxic arsenite [As (III)] to less toxic arsenate [As (V)] that is easily removed using chemical and biological trappers. In this study, genomic and metagenomic approaches parallel to culture-based assay (Graphical abstract) have made it possible to decipher phylogenetic diversity of groundwater arsenotrophic microbiomes along with elucidation of their genetic determinants.
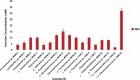
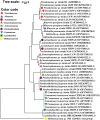
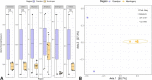
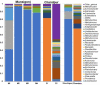
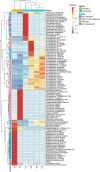
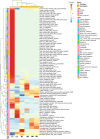
Results: Seventy-two isolates were retrieved from six As-contaminated (average As concentration of 0.23 mg/L) groundwater samples from Munshiganj and Chandpur districts of Bangladesh. Twenty-three isolates harbored arsenite efflux pump (arsB) gene with high abundance, and ten isolates possessing arsenite oxidase (aioA) gene, with a wide range of minimum inhibitory concentration, MICAs (2 to 32 mM), confirming their role in arsenite metabolism. There was considerable heterogeneity in species richness and microbial community structure. Microbial taxa from Proteobacteria, Firmicutes and Acidobacteria dominated these diversities. Through these combinatorial approaches, we have identified potential candidates such as, Pseudomonas, Acinetobacter, Stenotrophomonas, Achromobacter, Paraburkholderia, Comamonas and Klebsiella and associated functional genes (arsB, acr3, arsD, arsH, arsR) that could significantly contribute to arsenite detoxification, accumulation, and immobilization.
Conclusions: Culture-dependent and -independent shotgun metagenomic investigation elucidated arsenotrophic microbiomes and their functions in As biogeochemical transformation. These findings laid a foundation for further large-scale researches on the arsenotrophic microbiomes and their concurrent functions in As biogeochemical transformation in As-contaminated areas of Bangladesh and beyond.
Keywords: AMRs; Arsenotrophic bacteria; Arsenotrophic genes; Bioremediation; VFGs; Whole-genome shotgun sequencing.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Rahaman MS, Mise N, Ichihara S. Arsenic contamination in food chain in Bangladesh: a review on health hazards, socioeconomic impacts and implications. Hyg Environ Health Adv. 2022;2:100004.
-
- Hassan Z, Sultana M, van Breukelen BM, Khan SI, Röling WF. Diverse arsenic-and iron-cycling microbial communities in arsenic-contaminated aquifers used for drinking water in Bangladesh. FEMS Microbiol Ecol. 2015;91:fiv026. - PubMed
-
- Cavalca L, Corsini A, Zaccheo P, Andreoni V, Muyzer G. Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Future Microbiol. 2013;8:753–768. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

