Atl (atlastin) regulates mTor signaling and autophagy in Drosophila muscle through alteration of the lysosomal network
- PMID: 37649246
- PMCID: PMC10761077
- DOI: 10.1080/15548627.2023.2249794
Atl (atlastin) regulates mTor signaling and autophagy in Drosophila muscle through alteration of the lysosomal network
Abstract
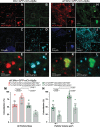
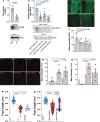
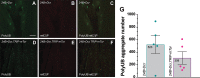
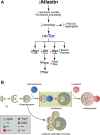
atl atlastin; ALR autophagic lysosome reformation; ER endoplasmic reticulum; GFP green fluorescent protein; HSP hereditary spastic paraplegia; Lamp1 lysosomal associated membrane protein 1 PolyUB polyubiquitin; RFP red fluorescent protein; spin spinster; mTor mechanistic Target of rapamycin; VCP valosin containing protein.
Keywords: ATG8; Hereditary Spastic Paraplegia; LAMP1; Neurodegeneration; endosome; protein aggregates; rab7.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


Similar articles
-
Mouse models for hereditary spastic paraplegia uncover a role of PI4K2A in autophagic lysosome reformation.Autophagy. 2021 Nov;17(11):3690-3706. doi: 10.1080/15548627.2021.1891848. Epub 2021 Mar 9. Autophagy. 2021. PMID: 33618608 Free PMC article.
-
Spinster is required for autophagic lysosome reformation and mTOR reactivation following starvation.Proc Natl Acad Sci U S A. 2011 May 10;108(19):7826-31. doi: 10.1073/pnas.1013800108. Epub 2011 Apr 25. Proc Natl Acad Sci U S A. 2011. PMID: 21518918 Free PMC article.
-
ZFYVE26/SPASTIZIN and SPG11/SPATACSIN mutations in hereditary spastic paraplegia types AR-SPG15 and AR-SPG11 have different effects on autophagy and endocytosis.Autophagy. 2019 Jan;15(1):34-57. doi: 10.1080/15548627.2018.1507438. Epub 2018 Sep 13. Autophagy. 2019. PMID: 30081747 Free PMC article.
-
Recent progress in autophagic lysosome reformation.Traffic. 2017 Jun;18(6):358-361. doi: 10.1111/tra.12484. Epub 2017 May 5. Traffic. 2017. PMID: 28371052 Review.
-
MTOR, PIK3C3, and autophagy: Signaling the beginning from the end.Autophagy. 2015;11(12):2375-6. doi: 10.1080/15548627.2015.1106668. Autophagy. 2015. PMID: 26565689 Free PMC article. Review.
Cited by
-
Atlastin-1 regulates endosomal tubulation and lysosomal proteolysis in human cortical neurons.Neurobiol Dis. 2024 Sep;199:106556. doi: 10.1016/j.nbd.2024.106556. Epub 2024 Jun 6. Neurobiol Dis. 2024. PMID: 38851544 Free PMC article.
-
Molecular mechanism on autophagy associated cardiovascular dysfunction in Drosophila melanogaster.Front Cell Dev Biol. 2025 Mar 3;13:1512341. doi: 10.3389/fcell.2025.1512341. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40099194 Free PMC article. Review.
-
Motor neuron activity enhances the proteomic stress caused by autophagy defects in the target muscle.PLoS One. 2024 Jan 2;19(1):e0291477. doi: 10.1371/journal.pone.0291477. eCollection 2024. PLoS One. 2024. PMID: 38166124 Free PMC article.
-
Loss of Fic causes progressive neurodegeneration in a Drosophila model of hereditary spastic paraplegia.Biochim Biophys Acta Mol Basis Dis. 2024 Oct;1870(7):167348. doi: 10.1016/j.bbadis.2024.167348. Epub 2024 Jul 8. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38986817
-
The Drosophila Nesprin-1 homolog MSP300 is required for muscle autophagy and proteostasis.J Cell Sci. 2024 Jun 1;137(11):jcs262096. doi: 10.1242/jcs.262096. Epub 2024 Jun 10. J Cell Sci. 2024. PMID: 38757366 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
