Evolution of T cell receptor beta loci in salmonids
- PMID: 37649482
- PMCID: PMC10464911
- DOI: 10.3389/fimmu.2023.1238321
Evolution of T cell receptor beta loci in salmonids
Abstract
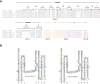
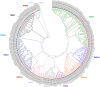
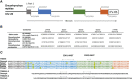
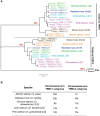
T-cell mediated immunity relies on a vast array of antigen specific T cell receptors (TR). Characterizing the structure of TR loci is essential to study the diversity and composition of T cell responses in vertebrate species. The lack of good-quality genome assemblies, and the difficulty to perform a reliably mapping of multiple highly similar TR sequences, have hindered the study of these loci in non-model organisms. High-quality genome assemblies are now available for the two main genera of Salmonids, Salmo and Oncorhynchus. We present here a full description and annotation of the TRB loci located on chromosomes 19 and 25 of rainbow trout (Oncorhynchus mykiss). To get insight about variations of the structure and composition of TRB locus across salmonids, we compared rainbow trout TRB loci with other salmonid species and confirmed that the basic structure of salmonid TRB locus is a double set of two TRBV-D-J-C loci in opposite orientation on two different chromosomes. Our data shed light on the evolution of TRB loci in Salmonids after their whole genome duplication (WGD). We established a coherent nomenclature of salmonid TRB loci based on comprehensive annotation. Our work provides a fundamental basis for monitoring salmonid T cell responses by TRB repertoire sequencing.
Keywords: T cell receptor; TRB locus; adaptive immunity; evolution; rainbow trout; repertoire; salmonid fish.
Copyright © 2023 Boudinot, Novas, Jouneau, Mondot, Lefranc, Grimholt and Magadán.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Flajnik M, Du Pasquier L. Evolution of the immune system. In: Paul W, editor. Fundam. Immunol., 7th edn. (2012) New York: Wolters Kluwer & Lippincott Willians & Wilkins. p. 67–128.
-
- Lefranc M-P, Lefranc G. The immunoglobulin FactsBook. London (UK): Academic Press; (2001).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

