Global analysis of HLA-A2 restricted MAGE-A3 tumor antigen epitopes and corresponding TCRs in non-small cell lung cancer
- PMID: 37649599
- PMCID: PMC10465222
- DOI: 10.7150/thno.84710
Global analysis of HLA-A2 restricted MAGE-A3 tumor antigen epitopes and corresponding TCRs in non-small cell lung cancer
Abstract
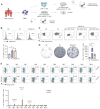
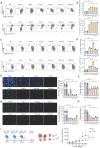
Background: Advanced non-small cell lung cancer (NSCLC) is the most common type of lung cancer with poor prognosis. Adoptive cell therapy using engineered T-cell receptors (TCRs) targeting cancer-testis antigens, such as Melanoma-associated antigen 3 (MAGE-A3), is a potential approach for the treatment of NSCLC. However, systematic analysis of T cell immune responses to MAGE-A3 antigen and corresponding antigen-specific TCR is still lacking. Methods: In this study, we comprehensively screened HLA-A2 restricted MAGE-A3 tumor epitopes and characterized the corresponding TCRs using in vitro artificial antigen presentation cells (APC) system, single-cell transcriptome and TCR V(D)J sequencing, and machine-learning. Furthermore, the tumor-reactive TCRs with killing potency was screened and verified. Results: We identified the HLA-A2 restricted T cell epitopes from MAGE-A3 that could effectively induce the activation and cytotoxicity of CD8+ T cells using artificial APC in vitro. A cohort of HLA-A2+ NSCLC donors demonstrated that the number of epitope specific CD8+ T cells increased in NSCLC than healthy controls when measured with tetramer derived from the candidate MAGE-A3 epitopes, especially epitope Mp4 (MAGE-A3: 160-169, LVFGIELMEV). Statistical and machine-learning based analyses demonstrated that the MAGE-A3-Mp4 epitope-specific CD8+ T cell clones were mostly in effector and proliferating state. Importantly, T cells artificially expressing the MAGE-A3-Mp4 specific TCRs exhibited strong MAGE-A3+ tumor cell recognition and killing effect. Cross-reactivity risk analysis of the candidates TCRs showed high binding stability to MAGE-A3-Mp4 epitope and low risk of cross-reaction. Conclusions: This work identified candidate TCRs potentially suitable for TCR-T design targeting HLA-A2 restricted MAGE-A3 tumor antigen.
Keywords: Cancer immunotherapy; Epitope-specific TCR; MAGE-A3; NSCLC.
© The author(s).
Conflict of interest statement
Competing Interests: Patent applications have been filed on aspects of the described candidate TCR sequences for TCR-T design by O.J.L., B.Z., Z.R., G.C., L.G. and H.Z. The other authors declare no competing interests.
Figures






References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A. et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535–54. - PubMed
-
- Planchard D, Popat S, Kerr K, Novello S, Smit EF, Faivre-Finn C. et al. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv192–iv237. - PubMed
-
- Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer. 2021;21:298–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

