Genomic and tumour microenvironmental biomarkers of immune checkpoint inhibitor response in advanced Taiwanese melanoma
- PMID: 37649975
- PMCID: PMC10463562
- DOI: 10.1002/cti2.1465
Genomic and tumour microenvironmental biomarkers of immune checkpoint inhibitor response in advanced Taiwanese melanoma
Abstract
Objective: Genomic biomarkers predicting immune checkpoint inhibitor (ICI) treatment outcomes for Asian metastatic melanoma have been rarely reported. This study presents data on next-generation sequencing (NGS) and tumour microenvironment biomarkers in 33 cases.
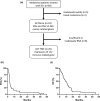
Methods: Thirty-three patients with advanced melanoma, who underwent ICI treatment at the Chang Gung Memorial Hospital in Taiwan, were recruited. The study evaluated clinical outcomes, including response rate, disease control rate, progression-free survival (PFS) rate and overall survival (OS) rate. Archived tissue samples from 33 cases were subjected to NGS by ACTOnco, and ACTTME was employed in 25 cases.
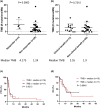
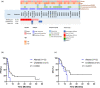
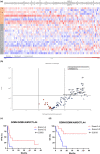
Results: The most prevalent driver mutations were BRAF mutations (24.2%), followed by NRAS (15.2%), KIT (12.1%), KRAS (9.1%) and NF1 (9.1%) mutations. Acral/mucosal melanomas exhibited distinct mutation patterns compared to non-acral melanomas. Tumour mutational burden estimated using ACTOnco was not associated with ICI efficacy. Notably, genetic alterations in the p53 pathway (CDKNA2 loss, MDM2 gain/amplification and TP53 mutation) accounted for 36.4% and were significantly associated with unfavourable PFS (median PFS 2.7 months vs. 3.9 months, P = 0.0394). Moreover, 26 genes were identified as differentially expressed genes that were upregulated in patients with clinical benefits compared to those without benefits. Four genes, GZMH, GZMK, AIM2 and CTLA4, were found to be associated with both PFS and OS.
Conclusion: Genetic alterations in the p53 pathway may be critical in Asian patients with melanoma undergoing ICI treatment. Further investigation is required to explore this mechanism and validate these findings.
Keywords: ICIs; genomic and TME biomarkers; melanoma; p53.
© 2023 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
KTT was an employee of ACT Genomics Co., Ltd. CLW, YHW and SJC are employees of ACT Genomics Co., Ltd. The other authors declare that the research was conducted without commercial or financial relationships that could be construed as potential conflicts of interest.
Figures

Similar articles
-
Biologic subtypes of melanoma predict survival benefit of combination anti-PD1+anti-CTLA4 immune checkpoint inhibitors versus anti-PD1 monotherapy.J Immunother Cancer. 2021 Jan;9(1):e001642. doi: 10.1136/jitc-2020-001642. J Immunother Cancer. 2021. PMID: 33483342 Free PMC article.
-
Distinct Mutation Patterns Reveal Melanoma Subtypes and Influence Immunotherapy Response in Advanced Melanoma Patients.Cancers (Basel). 2020 Aug 20;12(9):2359. doi: 10.3390/cancers12092359. Cancers (Basel). 2020. PMID: 32825510 Free PMC article.
-
Genetic characterization of advanced conjunctival melanoma and response to systemic treatment.Eur J Cancer. 2022 May;166:60-72. doi: 10.1016/j.ejca.2022.01.008. Epub 2022 Mar 10. Eur J Cancer. 2022. PMID: 35279471
-
Validation of Progression-Free Survival Rate at 6 Months and Objective Response for Estimating Overall Survival in Immune Checkpoint Inhibitor Trials: A Systematic Review and Meta-analysis.JAMA Netw Open. 2020 Sep 1;3(9):e2011809. doi: 10.1001/jamanetworkopen.2020.11809. JAMA Netw Open. 2020. PMID: 32897371 Free PMC article.
-
Comprehensive clinicopathological and molecular analysis of primary malignant melanoma of the oesophagus.Histopathology. 2021 Jan;78(2):240-251. doi: 10.1111/his.14210. Epub 2020 Sep 20. Histopathology. 2021. PMID: 32654197 Review.
Cited by
-
Pro-inflammatory granzyme K contributes extracellularly to disease.Front Immunol. 2025 Jun 18;16:1620670. doi: 10.3389/fimmu.2025.1620670. eCollection 2025. Front Immunol. 2025. PMID: 40607408 Free PMC article. Review.
-
Innate Immune Cells in Melanoma: Implications for Immunotherapy.Int J Mol Sci. 2024 Aug 5;25(15):8523. doi: 10.3390/ijms25158523. Int J Mol Sci. 2024. PMID: 39126091 Free PMC article. Review.
-
Genomic landscape and comparative analysis of tissue and liquid-based NGS in Taiwanese anaplastic thyroid carcinoma.NPJ Precis Oncol. 2025 Jan 14;9(1):16. doi: 10.1038/s41698-025-00802-2. NPJ Precis Oncol. 2025. PMID: 39809865 Free PMC article.
-
Granzyme K drives a newly-intentified pathway of complement activation.bioRxiv [Preprint]. 2024 May 26:2024.05.22.595315. doi: 10.1101/2024.05.22.595315. bioRxiv. 2024. Update in: Nature. 2025 May;641(8061):211-221. doi: 10.1038/s41586-025-08713-9. PMID: 38826230 Free PMC article. Updated. Preprint.
References
-
- Larkin J, Chiarion‐Sileni V, Gonzalez R et al. Five‐year survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2019; 381: 1535–1546. - PubMed
-
- Robert C, Ribas A, Schachter J et al. Pembrolizumab versus ipilimumab in advanced melanoma (KEYNOTE‐006): Post‐hoc 5‐year results from an open‐label, multicentre, randomised, controlled, phase 3 study. Lancet Oncol 2019; 20: 1239–1251. - PubMed
-
- Hayward NK, Wilmott JS, Waddell N et al. Whole‐genome landscapes of major melanoma subtypes. Nature 2017; 545: 175–180. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
