Variant-Specific Viral Kinetics in Acute COVID-19
- PMID: 37650233
- PMCID: PMC10469346
- DOI: 10.1093/infdis/jiad314
Variant-Specific Viral Kinetics in Acute COVID-19
Abstract
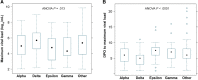
Understanding variant-specific differences in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral kinetics may explain differences in transmission efficiency and provide insights on pathogenesis and prevention. We evaluated SARS-CoV-2 kinetics from nasal swabs across multiple variants (Alpha, Delta, Epsilon, Gamma) in placebo recipients of the ACTIV-2/A5401 trial. Delta variant infection led to the highest maximum viral load and shortest time from symptom onset to viral load peak. There were no significant differences in time to viral clearance across the variants. Viral decline was biphasic with first- and second-phase decays having half-lives of 11 hours and 2.5 days, respectively, with differences among variants, especially in the second phase. These results suggest that while variant-specific differences in viral kinetics exist, post-peak viral load all variants appeared to be efficiently cleared by the host. Clinical Trials Registration. NCT04518410.
Keywords: COVID-19; variant; viral kinetics.
© The Author(s) 2023. Published by Oxford University Press on behalf of Infectious Diseases Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Conflict of interest statement
Potential conflicts of interest. K. W. C. received research funding to institution from Merck Sharp & Dohme, and was a consultant for Pardes Biosciences. J. Z. L. was a consultant for AbbVie and received research support from Merck. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Figures


References
Publication types
MeSH terms
Supplementary concepts
Associated data
Grants and funding
- UM1 AI069423/AI/NIAID NIH HHS/United States
- UM1 AI106701/AI/NIAID NIH HHS/United States
- UM1 AI069424/AI/NIAID NIH HHS/United States
- R01-HL143541);/NH/NIH HHS/United States
- U01 AI069463/AI/NIAID NIH HHS/United States
- UM1 AI069463/AI/NIAID NIH HHS/United States
- UM1 AI069412/AI/NIAID NIH HHS/United States
- UM1 AI068634/AI/NIAID NIH HHS/United States
- UM1 AI068636/AI/NIAID NIH HHS/United States
- U01 AI068636/AI/NIAID NIH HHS/United States
- U54 HL143541/HL/NHLBI NIH HHS/United States
- U01 AI069424/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

