A Machine Learning Analysis of Prognostic Genes Associated With Allograft Tolerance After Renal Transplantation
- PMID: 37650419
- PMCID: PMC10475226
- DOI: 10.1177/09636897231195116
A Machine Learning Analysis of Prognostic Genes Associated With Allograft Tolerance After Renal Transplantation
Abstract
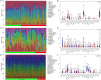
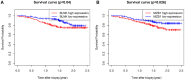
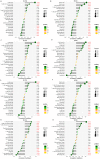
In this study, we aimed to identify transplantation tolerance (TOL)-related gene signature and use it to predict the different types of renal allograft rejection performances in kidney transplantation. Gene expression data were obtained from the Gene Expression Omnibus (GEO) database, differently expressed genes (DEGs) were performed, and the gene ontology (GO) function enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were also conducted. The machine learning methods were combined to analyze the feature TOL-related genes and verify their predictive performance. Afterward, the gene expression levels and predictive performances of TOL-related genes were conducted in the context of acute rejection (AR), chronic rejection (CR), and graft loss through heatmap plots and the receiver operating characteristic (ROC) curves, and their respective immune infiltration results were also performed. Furthermore, the TOL-related gene signature for graft survival was conducted to discover gene immune cell enrichment. A total of 25 TOL-related DEGs were founded, and the GO and KEGG results indicated that DEGs mainly enriched in B cell-related functions and pathways. 7 TOL-related gene signature was constructed and performed delightedly in TOL groups and different types of allograft rejection. The immune infiltration analysis suggested that gene signature was correlated with different types of immune cells. The Kaplan-Meier (KM) survival analysis demonstrated that BLNK and MZB1 were the prognostic TOL-related genes. Our study proposed a novel gene signature that may influence TOL in kidney transplantation, providing possible guidance for immunosuppressive therapy in kidney transplant patients.
Keywords: GEO; kidney transplantation; machine learning; transplantation tolerance.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures






Similar articles
-
Immune-Related Genes for Predicting Future Kidney Graft Loss: A Study Based on GEO Database.Front Immunol. 2022 Feb 25;13:859693. doi: 10.3389/fimmu.2022.859693. eCollection 2022. Front Immunol. 2022. PMID: 35281025 Free PMC article.
-
Identification of mitophagy-related gene signatures for predicting delayed graft function and renal allograft loss post-kidney transplantation.Transpl Immunol. 2024 Dec;87:102148. doi: 10.1016/j.trim.2024.102148. Epub 2024 Nov 14. Transpl Immunol. 2024. PMID: 39549926
-
Bioinformatics analysis characterizes immune infiltration landscape and identifies potential blood biomarkers for heart transplantation.Transpl Immunol. 2024 Jun;84:102036. doi: 10.1016/j.trim.2024.102036. Epub 2024 Mar 16. Transpl Immunol. 2024. PMID: 38499050
-
Clinical operational tolerance after kidney transplantation: a short literature review.Transplant Proc. 2008 Jul-Aug;40(6):1847-51. doi: 10.1016/j.transproceed.2008.05.013. Transplant Proc. 2008. PMID: 18675067 Review.
-
Matchmaking the B-cell signature of tolerance to regulatory B cells.Am J Transplant. 2011 Dec;11(12):2555-60. doi: 10.1111/j.1600-6143.2011.03773.x. Epub 2011 Oct 3. Am J Transplant. 2011. PMID: 21967001 Free PMC article. Review.
References
-
- Rodrigue JR, Kazley AS, Mandelbrot DA, Hays R, LaPointe Rudow D, Baliga P, American Society of Transplantation. Living donor kidney transplantation: overcoming disparities in live kidney donation in the US–recommendations from a consensus conference. Clin J Am Soc Nephrol. 2015;10(9):1687–95. - PMC - PubMed
-
- Nafar M, Farrokhi F, Hemati K, Pour-Reza-Gholi F, Firoozan A, Einollahi B. Plasmapheresis in the treatment of early acute kidney allograft dysfunction. Exp Clin Transplant. 2006;4(2):506–509. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

