Targeting TREM1 augments antitumor T cell immunity by inhibiting myeloid-derived suppressor cells and restraining anti-PD-1 resistance
- PMID: 37651197
- PMCID: PMC10617775
- DOI: 10.1172/JCI167951
Targeting TREM1 augments antitumor T cell immunity by inhibiting myeloid-derived suppressor cells and restraining anti-PD-1 resistance
Abstract
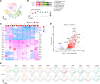
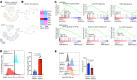
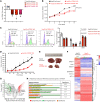
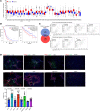
The triggering receptor expressed on myeloid cell 1 (TREM1) plays a critical role in development of chronic inflammatory disorders and the inflamed tumor microenvironment (TME) associated with most solid tumors. We examined whether loss of TREM1 signaling can abrogate the immunosuppressive TME and enhance cancer immunity. To investigate the therapeutic potential of TREM1 in cancer, we used mice deficient in Trem1 and developed a novel small molecule TREM1 inhibitor, VJDT. We demonstrated that genetic or pharmacological TREM1 silencing significantly delayed tumor growth in murine melanoma (B16F10) and fibrosarcoma (MCA205) models. Single-cell RNA-Seq combined with functional assays during TREM1 deficiency revealed decreased immunosuppressive capacity of myeloid-derived suppressor cells (MDSCs) accompanied by expansion in cytotoxic CD8+ T cells and increased PD-1 expression. Furthermore, TREM1 inhibition enhanced the antitumorigenic effect of anti-PD-1 treatment, in part, by limiting MDSC frequency and abrogating T cell exhaustion. In patient-derived melanoma xenograft tumors, treatment with VJDT downregulated key oncogenic signaling pathways involved in cell proliferation, migration, and survival. Our work highlights the role of TREM1 in cancer progression, both intrinsically expressed in cancer cells and extrinsically in the TME. Thus, targeting TREM1 to modify an immunosuppressive TME and improve efficacy of immune checkpoint therapy represents what we believe to be a promising therapeutic approach to cancer.
Keywords: Cancer immunotherapy; Cellular immune response; Melanoma; Oncology; Therapeutics.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

