A single inactivating amino acid change in the SARS-CoV-2 NSP3 Mac1 domain attenuates viral replication in vivo
- PMID: 37651466
- PMCID: PMC10499221
- DOI: 10.1371/journal.ppat.1011614
A single inactivating amino acid change in the SARS-CoV-2 NSP3 Mac1 domain attenuates viral replication in vivo
Abstract
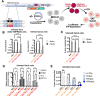
Despite unprecedented efforts, our therapeutic arsenal against SARS-CoV-2 remains limited. The conserved macrodomain 1 (Mac1) in NSP3 is an enzyme exhibiting ADP-ribosylhydrolase activity and a possible drug target. To determine the role of Mac1 catalytic activity in viral replication, we generated recombinant viruses and replicons encoding a catalytically inactive NSP3 Mac1 domain by mutating a critical asparagine in the active site. While substitution to alanine (N40A) reduced catalytic activity by ~10-fold, mutations to aspartic acid (N40D) reduced activity by ~100-fold relative to wild-type. Importantly, the N40A mutation rendered Mac1 unstable in vitro and lowered expression levels in bacterial and mammalian cells. When incorporated into SARS-CoV-2 molecular clones, the N40D mutant only modestly affected viral fitness in immortalized cell lines, but reduced viral replication in human airway organoids by 10-fold. In mice, the N40D mutant replicated at >1000-fold lower levels compared to the wild-type virus while inducing a robust interferon response; all animals infected with the mutant virus survived infection. Our data validate the critical role of SARS-CoV-2 NSP3 Mac1 catalytic activity in viral replication and as a promising therapeutic target to develop antivirals.
Copyright: © 2023 Taha et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
T.Y.T. and M.O. are inventors on a patent application filed by the Gladstone Institutes that covers the use of pGLUE to generate SARS-CoV-2 infectious clones and replicons. A.A. is a co-founder of Tango Therapeutics, Azkarra Therapeutics, Ovibio Corporation and Kytarro, a member of the board of Cytomx and Cambridge Science Corporation, a member of the scientific advisory board of Genentech, GLAdiator, Circle, Bluestar, Earli, Ambagon, Phoenix Molecular Designs and Trial Library, a consultant for SPARC, ProLynx, GSK and Novartis, receives grant or research support from SPARC and AstraZeneca, and holds patents on the use of PARP inhibitors held jointly with AstraZeneca from which he has benefited financially (and may do so in the future). The Krogan Laboratory has received research support from Vir Biotechnology, F. Hoffmann-La Roche, and Rezo Therapeutics. N.J.K has financially compensated consulting agreements with Maze Therapeutics, Interline Therapeutics, Rezo Therapeutics, and GEn1E Lifesciences, Inc.. N.J.K is on the Board of Directors of Rezo Therapeutics and is a shareholder in Tenaya Therapeutics, Maze Therapeutics, Rezo Therapeutics, and Interline Therapeutics. All other authors declare no competing interests.
Figures


Update of
-
A single inactivating amino acid change in the SARS-CoV-2 NSP3 Mac1 domain attenuates viral replication and pathogenesis in vivo.bioRxiv [Preprint]. 2023 May 10:2023.04.18.537104. doi: 10.1101/2023.04.18.537104. bioRxiv. 2023. Update in: PLoS Pathog. 2023 Aug 31;19(8):e1011614. doi: 10.1371/journal.ppat.1011614. PMID: 37131711 Free PMC article. Updated. Preprint.
References
-
- WHO. WHO Coronavirus (COVID-19) Dashboard: World Health Organization; 2022. [cited 2022 9/17/2022]. Available from: https://covid19.who.int/.
-
- Jayk Bernal A, Gomes da Silva MM, Musungaie DB, Kovalchuk E, Gonzalez A, Delos Reyes V, et al.. Molnupiravir for Oral Treatment of Covid-19 in Nonhospitalized Patients. N Engl J Med. 2022;386(6):509–20. Epub 2021/12/17. doi: 10.1056/NEJMoa2116044 ; PubMed Central PMCID: PMC8693688. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

