MpbPPI: a multi-task pre-training-based equivariant approach for the prediction of the effect of amino acid mutations on protein-protein interactions
- PMID: 37651610
- PMCID: PMC10516393
- DOI: 10.1093/bib/bbad310
MpbPPI: a multi-task pre-training-based equivariant approach for the prediction of the effect of amino acid mutations on protein-protein interactions
Abstract
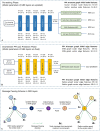
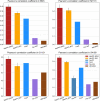
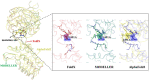
The accurate prediction of the effect of amino acid mutations for protein-protein interactions (PPI $\Delta \Delta G$) is a crucial task in protein engineering, as it provides insight into the relevant biological processes underpinning protein binding and provides a basis for further drug discovery. In this study, we propose MpbPPI, a novel multi-task pre-training-based geometric equivariance-preserving framework to predict PPI $\Delta \Delta G$. Pre-training on a strictly screened pre-training dataset is employed to address the scarcity of protein-protein complex structures annotated with PPI $\Delta \Delta G$ values. MpbPPI employs a multi-task pre-training technique, forcing the framework to learn comprehensive backbone and side chain geometric regulations of protein-protein complexes at different scales. After pre-training, MpbPPI can generate high-quality representations capturing the effective geometric characteristics of labeled protein-protein complexes for downstream $\Delta \Delta G$ predictions. MpbPPI serves as a scalable framework supporting different sources of mutant-type (MT) protein-protein complexes for flexible application. Experimental results on four benchmark datasets demonstrate that MpbPPI is a state-of-the-art framework for PPI $\Delta \Delta G$ predictions. The data and source code are available at https://github.com/arantir123/MpbPPI.
Keywords: equivariant neural network; multi-task pre-training; protein binding affinity change prediction; protein engineering.
© The Author(s) 2023. Published by Oxford University Press.
Figures

References
-
- Braun P, Gingras AC. History of protein–protein interactions: from egg-white to complex networks. Proteomics 2012;12(10):1478–98. - PubMed

