Finding significance: New perspectives in variant classification of the RAD51 regulators, BRCA2 and beyond
- PMID: 37651978
- PMCID: PMC10529980
- DOI: 10.1016/j.dnarep.2023.103563
Finding significance: New perspectives in variant classification of the RAD51 regulators, BRCA2 and beyond
Abstract
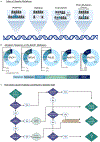
For many individuals harboring a variant of uncertain functional significance (VUS) in a homologous recombination (HR) gene, their risk of developing breast and ovarian cancer is unknown. Integral to the process of HR are BRCA1 and regulators of the central HR protein, RAD51, including BRCA2, PALB2, RAD51C and RAD51D. Due to advancements in sequencing technology and the continued expansion of cancer screening panels, the number of VUS identified in these genes has risen significantly. Standard practices for variant classification utilize different types of predictive, population, phenotypic, allelic and functional evidence. While variant analysis is improving, there remains a struggle to keep up with demand. Understanding the effects of an HR variant can aid in preventative care and is critical for developing an effective cancer treatment plan. In this review, we discuss current perspectives in the classification of variants in the breast and ovarian cancer genes BRCA1, BRCA2, PALB2, RAD51C and RAD51D.
Keywords: BRCA1; BRCA2; Breast cancer; Homologous recombination; Homologous recombination deficient tumors; Ovarian cancer; PALB2; RAD51; RAD51C; RAD51D; Variant of unknown/uncertain significance.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known conflicts of interest that would influence the contents of this manuscript.
Figures



References
-
- Varol U, Kucukzeybek Y, Alacacioglu A, Somali I, Altun Z, Aktas S, Oktay Tarhan M, BRCA genes: BRCA 1 and BRCA 2, J BUON, 23 (2018) 862–866. - PubMed
-
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. , A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1, Science, 266 (1994) 66–71. - PubMed
-
- Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, Huey B, King MC, Linkage of early-onset familial breast cancer to chromosome 17q21, Science, 250 (1990) 1684–1689. - PubMed
-
- Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. , Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13, Science, 265 (1994) 2088–2090. - PubMed
-
- Fanale D, Pivetti A, Cancelliere D, Spera A, Bono M, Fiorino A, Pedone E, Barraco N, Brando C, Perez A, Guarneri MF, Russo TDB, Vieni S, Guarneri G, Russo A, Bazan V, BRCA1/2 variants of unknown significance in hereditary breast and ovarian cancer (HBOC) syndrome: Looking for the hidden meaning, Crit Rev Oncol Hematol, 172 (2022) 103626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

