Population serum proteomics uncovers a prognostic protein classifier for metabolic syndrome
- PMID: 37652016
- PMCID: PMC10518601
- DOI: 10.1016/j.xcrm.2023.101172
Population serum proteomics uncovers a prognostic protein classifier for metabolic syndrome
Abstract
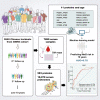
Metabolic syndrome (MetS) is a complex metabolic disorder with a global prevalence of 20%-25%. Early identification and intervention would help minimize the global burden on healthcare systems. Here, we measured over 400 proteins from ∼20,000 proteomes using data-independent acquisition mass spectrometry for 7,890 serum samples from a longitudinal cohort of 3,840 participants with two follow-up time points over 10 years. We then built a machine-learning model for predicting the risk of developing MetS within 10 years. Our model, composed of 11 proteins and the age of the individuals, achieved an area under the curve of 0.774 in the validation cohort (n = 242). Using linear mixed models, we found that apolipoproteins, immune-related proteins, and coagulation-related proteins best correlated with MetS development. This population-scale proteomics study broadens our understanding of MetS and may guide the development of prevention and targeted therapies for MetS.
Keywords: DIA-MS; apolipoproteins; blood proteomics; diabetes; high-density lipoproteins; machine learning; metabolic syndrome; population proteomics; prospective prediction; triglycerides.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests Y. Zhu and T.G. are shareholders of Westlake Omics, Inc. B.W., W.G., and N.X. are employees of Westlake Omics, Inc. A patent related to this study has been submitted to the China National Intellectual Property Administration.
Figures






References
-
- Expert Panel on Detection, E., and Treatment of High Blood Cholesterol in, Expert Panel on Detection Evaluation and Treatment of High Blood Cholesterol in Adults Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) JAMA. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. - DOI - PubMed
-
- International Diabetes Federation: The IDF consensus worldwide definition of the metabolic syndrome.
-
- do Vale Moreira N.C., Hussain A., Bhowmik B., Mdala I., Siddiquee T., Fernandes V.O., Montenegro Júnior R.M., Meyer H.E. Prevalence of Metabolic Syndrome by different definitions, and its association with type 2 diabetes, pre-diabetes, and cardiovascular disease risk in Brazil. Diabetes Metabol. Syndr. 2020;14:1217–1224. doi: 10.1016/j.dsx.2020.05.043. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

