IL-13-induced STAT3-dependent signaling networks regulate esophageal epithelial proliferation in eosinophilic esophagitis
- PMID: 37652141
- PMCID: PMC11102758
- DOI: 10.1016/j.jaci.2023.07.021
IL-13-induced STAT3-dependent signaling networks regulate esophageal epithelial proliferation in eosinophilic esophagitis
Abstract
Background: Basal zone hyperplasia (BZH) and dilated intercellular spaces (DISs) are thought to contribute to the clinical manifestations of eosinophilic esophagitis (EoE); however, the molecular pathways that drive BZH remain largely unexplored.
Objective: We sought to define the role of IL-13-induced transcriptional programs in esophageal epithelial proliferation in EoE.
Methods: We performed RNA sequencing, bioinformatics, Western blot, reverse transcriptase quantitative PCR, and histologic analyses on esophageal biopsies from healthy control and patients with EoE, primary esophageal cells derived from patients with EoE, and IL-13-stimulated esophageal epithelial keratinocytes grown at the air-liquid interface (EPC2-ALI). Genetic (shRNA) and pharmacologic (proteolysis-targeting chimera degrader) approaches and in vivo model of IL-13-induced esophageal epithelial remodeling (Krt5-rtTA x tetO-IL-13Tg) were used to define the role of signal transducer and activator of transcription 3 (STAT3) and STAT6 and secreted frizzled-related protein 1 (SFRP1) in esophageal epithelial proliferation.
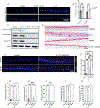
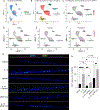
Results: RNA-sequencing analysis of esophageal biopsies (healthy control vs EoE) and EPC2-ALI revealed 82 common differentially expressed genes that were enriched for putative STAT3 target genes. In vitro and in vivo analyses revealed a link between IL-13-induced STAT3 and STAT6 phosphorylation, SFRP1 mRNA expression, and esophageal epithelial proliferation. In vitro studies showed that IL-13-induced esophageal epithelial proliferation was STAT3-dependent and regulated by the STAT3 target SFRP1. SFRP1 mRNA is increased in esophageal biopsies from patients with active EoE compared with healthy controls or patients in remission and identifies an esophageal suprabasal epithelial cell subpopulation that uniquely expressed the core EoE proinflammatory transcriptome genes (CCL26, ALOX15, CAPN14, ANO1, and TNFAIP6).
Conclusions: These studies identify SFRP1 as a key regulator of IL-13-induced and STAT3-dependent esophageal proliferation and BZH in EoE and link SFRP1+ esophageal epithelial cells with the proinflammatory and epithelial remodeling response in EoE.
Keywords: Eosinophilic esophagitis, epithelium, proliferation, basal zone hyperplasia, STAT3, SFRP1, bioinformatics.
Copyright © 2023 American Academy of Allergy, Asthma & Immunology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures






References
-
- Collins MH. Histopathology of eosinophilic esophagitis. Dig Dis 2014; 32:68–73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

