Screening non-conventional yeasts for acid tolerance and engineering Pichia occidentalis for production of muconic acid
- PMID: 37652930
- PMCID: PMC10471774
- DOI: 10.1038/s41467-023-41064-5
Screening non-conventional yeasts for acid tolerance and engineering Pichia occidentalis for production of muconic acid
Abstract
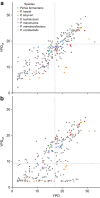
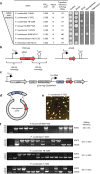
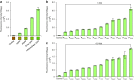
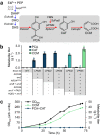
Saccharomyces cerevisiae is a workhorse of industrial biotechnology owing to the organism's prominence in alcohol fermentation and the suite of sophisticated genetic tools available to manipulate its metabolism. However, S. cerevisiae is not suited to overproduce many bulk bioproducts, as toxicity constrains production at high titers. Here, we employ a high-throughput assay to screen 108 publicly accessible yeast strains for tolerance to 20 g L-1 adipic acid (AA), a nylon precursor. We identify 15 tolerant yeasts and select Pichia occidentalis for production of cis,cis-muconic acid (CCM), the precursor to AA. By developing a genome editing toolkit for P. occidentalis, we demonstrate fed-batch production of CCM with a maximum titer (38.8 g L-1), yield (0.134 g g-1 glucose) and productivity (0.511 g L-1 h-1) that surpasses all metrics achieved using S. cerevisiae. This work brings us closer to the industrial bioproduction of AA and underscores the importance of host selection in bioprocessing.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Knoshaug EP, Zhang M. Butanol tolerance in a selection of microorganisms. Appl. Biochem. Biotechnol. 2009;153:13–20. - PubMed
-
- Long A, Ward O. Biotransformation of benzaldehyde by Saccharomyces cerevisiae: characterization of the fermentation and toxicity effects of substrates and products. Biotechnol. Bioeng. 1989;34:933–941. - PubMed
-
- Narendranath N, Thomas K, Ingledew W. Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J. Ind. Microbiol. Biotechnol. 2001;26:171–177. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

