CircFam190a: a critical positive regulator of osteoclast differentiation via enhancement of the AKT1/HSP90β complex
- PMID: 37653038
- PMCID: PMC10545668
- DOI: 10.1038/s12276-023-01085-y
CircFam190a: a critical positive regulator of osteoclast differentiation via enhancement of the AKT1/HSP90β complex
Abstract
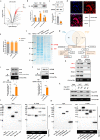
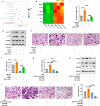
The identification of key regulatory factors that control osteoclastogenesis is important. Accumulating evidence indicates that circular RNAs (circRNAs) are discrete functional entities. However, the complexities of circRNA expression as well as the extent of their regulatory functions during osteoclastogenesis have yet to be revealed. Here, based on circular RNA sequencing data, we identified a circular RNA, circFam190a, as a critical regulator of osteoclast differentiation and function. During osteoclastogenesis, circFam190a is significantly upregulated. In vitro, circFam190a enhanced osteoclast formation and function. In vivo, overexpression of circFam190a induced significant bone loss, while knockdown of circFam190a prevented pathological bone loss in an ovariectomized (OVX) mouse osteoporosis model. Mechanistically, our data suggest that circFam90a enhances the binding of AKT1 and HSP90β, promoting AKT1 stability. Altogether, our findings highlight the critical role of circFam190a as a positive regulator of osteoclastogenesis, and targeting circFam190a might be a promising therapeutic strategy for treating pathological bone loss.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

