Mechanisms of antigen escape from BCMA- or GPRC5D-targeted immunotherapies in multiple myeloma
- PMID: 37653344
- PMCID: PMC10504087
- DOI: 10.1038/s41591-023-02491-5
Mechanisms of antigen escape from BCMA- or GPRC5D-targeted immunotherapies in multiple myeloma
Abstract
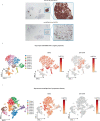
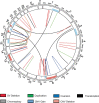
B cell maturation antigen (BCMA) target loss is considered to be a rare event that mediates multiple myeloma (MM) resistance to anti-BCMA chimeric antigen receptor T cell (CAR T) or bispecific T cell engager (TCE) therapies. Emerging data report that downregulation of G-protein-coupled receptor family C group 5 member D (GPRC5D) protein often occurs at relapse after anti-GPRC5D CAR T therapy. To examine the tumor-intrinsic factors that promote MM antigen escape, we performed combined bulk and single-cell whole-genome sequencing and copy number variation analysis of 30 patients treated with anti-BCMA and/or anti-GPRC5D CAR T/TCE therapy. In two cases, MM relapse post-TCE/CAR T therapy was driven by BCMA-negative clones harboring focal biallelic deletions at the TNFRSF17 locus at relapse or by selective expansion of pre-existing subclones with biallelic TNFRSF17 loss. In another five cases of relapse, newly detected, nontruncating, missense mutations or in-frame deletions in the extracellular domain of BCMA negated the efficacies of anti-BCMA TCE therapies, despite detectable surface BCMA protein expression. In the present study, we also report four cases of MM relapse with biallelic mutations of GPRC5D after anti-GPRC5D TCE therapy, including two cases with convergent evolution where multiple subclones lost GPRC5D through somatic events. Immunoselection of BCMA- or GPRC5D-negative or mutant clones is an important tumor-intrinsic driver of relapse post-targeted therapies. Mutational events on BCMA confer distinct sensitivities toward different anti-BCMA therapies, underscoring the importance of considering the tumor antigen landscape for optimal design and selection of targeted immunotherapies in MM.
© 2023. The Author(s).
Conflict of interest statement
N.J.B. has received research funding from Pfizer and speaker’s bureau honoraria from Amgen, BMS, Sanofi, Pfizer and Janssen; he is a consultant/advisory board member for BMS, Janssen and Pfizer. P.N. received speaker’s bureau honoraria from BMS, Janssen and Sanofi, and is a consultant/advisory board member for BMS and Janssen. C.H. and T.H. have equity ownership of MLL Munich Leukemia Laboratory. O.L. has received research funding from: the National Institutes of Health (NIH), NCI, US Food and Drug Administration, MMRF, International Myeloma Foundation, Leukemia and Lymphoma Society, the Paula and Rodger Riney Myeloma Foundation, Perelman Family Foundation, Rising Tide Foundation, Amgen, Celgene, Janssen, Takeda, Glenmark, Seattle Genetics and Karyopharm; received honoraria and is on advisory boards for Adaptive, Amgen, Binding Site, BMS, Celgene, Cellectis, Glenmark, Janssen, Juno and Pfizer; and serves on independent data monitoring committees for clinical trials led by Takeda, Merck, Janssen and Theradex. J.J.K has received research funding from Amgen, Genentech and Janssen, and has received honoraria and is on advisory boards for Janssen. The other authors declare no competing interests.
Figures














Comment in
-
Decoding antigen escape and multiple myeloma relapse after targeted immunotherapy.Nat Med. 2023 Sep;29(9):2185-2186. doi: 10.1038/s41591-023-02515-0. Nat Med. 2023. PMID: 37684544 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

