Immune-related risk prognostic model for clear cell renal cell carcinoma: Implications for immunotherapy
- PMID: 37653791
- PMCID: PMC10470711
- DOI: 10.1097/MD.0000000000034786
Immune-related risk prognostic model for clear cell renal cell carcinoma: Implications for immunotherapy
Abstract
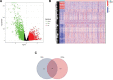
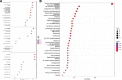
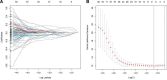
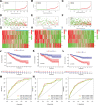
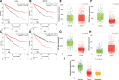
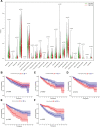
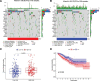
Clear cell renal cell carcinoma (ccRCC) is associated with complex immune interactions. We conducted a comprehensive analysis of immune-related differentially expressed genes in patients with ccRCC using data from The Cancer Genome Atlas and ImmPort databases. The immune-related differentially expressed genes underwent functional and pathway enrichment analysis, followed by COX regression combined with LASSO regression to construct an immune-related risk prognostic model. The model comprised 4 IRGs: CLDN4, SEMA3G, CAT, and UCN. Patients were stratified into high-risk and low-risk groups based on the median risk score, and the overall survival rate of the high-risk group was significantly lower than that of the low-risk group, confirming the reliability of the model from various perspectives. Further comparison of immune infiltration, tumor mutation load, and immunophenoscore (IPS) comparison between the 2 groups indicates that the high-risk group could potentially demonstrate a heightened sensitivity towards immunotherapy checkpoints PD-1, CTLA-4, IL-6, and LAG3 in ccRCC patients. The proposed model not only applies to ccRCC but also shows potential in developing into a prognostic model for renal cancer, thus introducing a novel approach for personalized immunotherapy in ccRCC.
Copyright © 2023 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures


References
-
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Motzer RJ, Bacik J, Mazumdar M. Prognostic factors for survival of patients with stage IV renal cell carcinoma: memorial sloan-kettering cancer center experience. Clin Cancer Res. 2004;10:6302S–3S. - PubMed
-
- Wettersten HI, Abu Aboud O, Lara PN, et al. Metabolic reprogramming in clear cell renal cell carcinoma. Nat Rev Nephrol. 2017;13:410–9. - PubMed
-
- Auvray M, Auclin E, Barthélémy P, et al. Second-line targeted therapies after nivolumab-ipilimumab failure in metastatic renal cell carcinoma. Eur J Cancer. 2019;108:33–40. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

