Development of lateral flow assays to detect host proteins in cattle for improved diagnosis of bovine tuberculosis
- PMID: 37655261
- PMCID: PMC10465798
- DOI: 10.3389/fvets.2023.1193332
Development of lateral flow assays to detect host proteins in cattle for improved diagnosis of bovine tuberculosis
Abstract
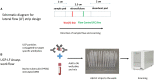
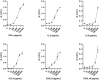
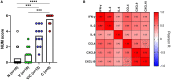
Bovine tuberculosis (bTB), caused by Mycobacterium bovis (M. bovis) infection in cattle, is an economically devastating chronic disease for livestock worldwide. Efficient disease control measures rely on early and accurate diagnosis using the tuberculin skin test (TST) and interferon-gamma release assays (IGRAs), followed by culling of positive animals. Compromised performance of TST and IGRA, due to BCG vaccination or co-infections with non-tuberculous mycobacteria (NTM), urges improved diagnostics. Lateral flow assays (LFAs) utilizing luminescent upconverting reporter particles (UCP) for quantitative measurement of host biomarkers present an accurate but less equipment- and labor-demanding diagnostic test platform. UCP-LFAs have proven applications for human infectious diseases. Here, we report the development of UCP-LFAs for the detection of six bovine proteins (IFN-γ, IL-2, IL-6, CCL4, CXCL9, and CXCL10), which have been described by ELISA as potential biomarkers to discriminate M. bovis infected from naïve and BCG-vaccinated cattle. We show that, in line with the ELISA data, the combined PPDb-induced levels of IFN-γ, IL-2, IL-6, CCL4, and CXCL9 determined by UCP-LFAs can discriminate M. bovis challenged animals from naïve (AUC range: 0.87-1.00) and BCG-vaccinated animals (AUC range: 0.97-1.00) in this cohort. These initial findings can be used to develop a robust and user-friendly multi-biomarker test (MBT) for bTB diagnosis.
Keywords: DIVA; UCP-LFA; biomarkers; bovine tuberculosis; chemokines; cytokines; diagnostics; upconverting reporter particles.
Copyright © 2023 Khalid, Pierneef, van Hooij, Zhou, de Jong, Tjon Kon Fat, Connelley, Hope, Corstjens and Geluk.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Correia CN, McHugo GP, Browne JA, McLoughlin KE, Nalpas NC, Magee DA, et al. . High-resolution transcriptomics of bovine purified protein derivative-stimulated peripheral blood from cattle infected with Mycobacterium bovis across an experimental time course. Tuberculosis. (2022) 136:102235. 10.1016/j.tube.2022.102235 - DOI - PubMed
-
- Refaya AK, Bhargavi G, Mathew NC, Rajendran A, Krishnamoorthy R, Swaminathan S, et al. . Bovine tuberculosis. Oxfordshire: CABI; (2018). - PubMed
-
- Palmer MV. Tuberculosis: a reemerging disease at the interface of domestic animals and wildlife. In: Childs JF, Mackenzie S, Richt JA, editors. Wildlife and Emerging Zoonotic Diseases: The Biology, Circumstances and Consequences of Cross-species Transmission. Berlin: Springer; (2007), p. 195–215. 10.1007/978-3-540-70962-6_9 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

