Immune checkpoint blockade induces distinct alterations in the microenvironments of primary and metastatic brain tumors
- PMID: 37655659
- PMCID: PMC10471177
- DOI: 10.1172/JCI169314
Immune checkpoint blockade induces distinct alterations in the microenvironments of primary and metastatic brain tumors
Abstract
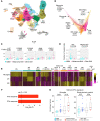
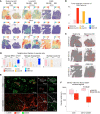
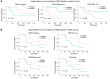
In comparison with responses in recurrent glioblastoma (rGBM), the intracranial response of brain metastases (BrM) to immune checkpoint blockade (ICB) is less well studied. Here, we present an integrated single-cell RNA-Seq (scRNA-Seq) study of 19 ICB-naive and 9 ICB-treated BrM samples from our own and published data sets. We compared them with our previously published scRNA-Seq data from rGBM and found that ICB led to more prominent T cell infiltration into BrM than rGBM. These BrM-infiltrating T cells exhibited a tumor-specific phenotype and displayed greater activated/exhausted features. We also used multiplex immunofluorescence and spatial transcriptomics to reveal that ICB reduced a distinct CD206+ macrophage population in the perivascular space, which may modulate T cell entry into BrM. Furthermore, we identified a subset of progenitor exhausted T cells that correlated with longer overall survival in BrM patients. Our study provides a comprehensive immune cellular landscape of ICB's effect on metastatic brain tumors and offers insights into potential strategies for improving ICB efficacy for brain tumor patients.
Keywords: Brain cancer; Cancer immunotherapy; Immunology; Neuroscience.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

