Mechanistic insights into sulfur source-driven physiological responses and metabolic reorganization in the fuel-biodesulfurizing Rhodococcus qingshengii IGTS8
- PMID: 37655899
- PMCID: PMC10537767
- DOI: 10.1128/aem.00826-23
Mechanistic insights into sulfur source-driven physiological responses and metabolic reorganization in the fuel-biodesulfurizing Rhodococcus qingshengii IGTS8
Abstract
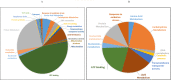
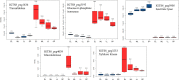
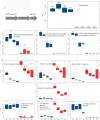
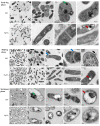
Comparative proteomics and untargeted metabolomics were combined to study the physiological and metabolic adaptations of Rhodococcus qingshengii IGTS8 under biodesulfurization conditions. After growth in a chemically defined medium with either dibenzothiophene (DBT) or MgSO4 as the sulfur source, many differentially produced proteins and metabolites associated with several metabolic and physiological processes were detected including the metabolism of carbohydrates, amino acids, lipids, nucleotides, vitamins, protein synthesis, transcriptional regulation, cell envelope biogenesis, and cell division. Increased production of the redox cofactor mycofactocin and associated proteins was one of the most striking adaptations under biodesulfurization conditions. While most central metabolic enzymes were less abundant in the presence of DBT, a key enzyme of the glyoxylate shunt, isocitrate lyase, was up to 26-fold more abundant. Several C1 metabolism and oligotrophy-related enzymes were significantly more abundant in the biodesulfurizing culture. R. qingshengii IGTS8 exhibited oligotrophic growth in liquid and solid media under carbon starvation. Moreover, the oligotrophic growth was faster on the solid medium in the presence of DBT compared to MgSO4 cultures. In the DBT culture, the cell envelope and phospholipids were remodeled, with lower levels of phosphatidylethanolamine and unsaturated and short-chain fatty acids being the most prominent changes. Biodesulfurization increased the biosynthesis of osmoprotectants (ectoine and mannosylglycerate) as well as glutamate and induced the stringent response. Our findings reveal highly diverse and overlapping stress responses that could protect the biodesulfurizing culture not only from the associated sulfate limitation but also from chemical, oxidative, and osmotic stress, allowing efficient resource management. IMPORTANCE Despite decades of research, a commercially viable bioprocess for fuel desulfurization has not been developed yet. This is mainly due to lack of knowledge of the physiology and metabolism of fuel-biodesulfurizing bacteria. Being a stressful condition, biodesulfurization could provoke several stress responses that are not understood. This is particularly important because a thorough understanding of the microbial stress response is essential for the development of environmentally friendly and industrially efficient microbial biocatalysts. Our comparative systems biology studies provide a mechanistic understanding of the biology of biodesulfurization, which is crucial for informed developments through the rational design of recombinant biodesulfurizers and optimization of the bioprocess conditions. Our findings enhance the understanding of the physiology, metabolism, and stress response not only in biodesulfurizing bacteria but also in rhodococci, a precious group of biotechnologically important bacteria.
Keywords: Rhodococcus; comparative proteomics; lipid metabolism; metabolomics; mycofactocin; oligotrophy; stress response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Kilbane JJ. 1990. Sulfur-specific microbial metabolism of organic compounds. Resour Conserv Recycl 3:69–79. doi: 10.1016/0921-3449(90)90046-7 - DOI
-
- Kilbane JJ, Bielaga B. 1990. Toward sulfur-free fuels. Chemtech 20:747–751.
Grants and funding
LinkOut - more resources
Full Text Sources

