System-wide mapping of peptide-GPCR interactions in C. elegans
- PMID: 37656621
- PMCID: PMC7615250
- DOI: 10.1016/j.celrep.2023.113058
System-wide mapping of peptide-GPCR interactions in C. elegans
Abstract
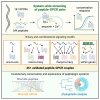
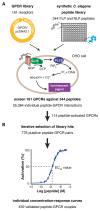
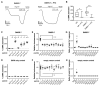
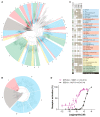
Neuropeptides and peptide hormones are ancient, widespread signaling molecules that underpin almost all brain functions. They constitute a broad ligand-receptor network, mainly by binding to G protein-coupled receptors (GPCRs). However, the organization of the peptidergic network and roles of many peptides remain elusive, as our insight into peptide-receptor interactions is limited and many peptide GPCRs are still orphan receptors. Here we report a genome-wide peptide-GPCR interaction map in Caenorhabditis elegans. By reverse pharmacology screening of over 55,384 possible interactions, we identify 461 cognate peptide-GPCR couples that uncover a broad signaling network with specific and complex combinatorial interactions encoded across and within single peptidergic genes. These interactions provide insights into peptide functions and evolution. Combining our dataset with phylogenetic analysis supports peptide-receptor co-evolution and conservation of at least 14 bilaterian peptidergic systems in C. elegans. This resource lays a foundation for system-wide analysis of the peptidergic network.
Keywords: C. elegans; CP: Cell biology; CP: Neuroscience; GPCR; interactome; neuropeptide; reverse pharmacology.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

