Comparative analysis of RNA secondary structure accuracy on predicted RNA 3D models
- PMID: 37656749
- PMCID: PMC10473517
- DOI: 10.1371/journal.pone.0290907
Comparative analysis of RNA secondary structure accuracy on predicted RNA 3D models
Abstract
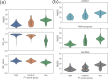
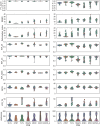
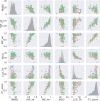
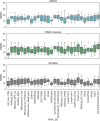
RNA structure is conformationally dynamic, and accurate all-atom tertiary (3D) structure modeling of RNA remains challenging with the prevailing tools. Secondary structure (2D) information is the standard prerequisite for most RNA 3D modeling. Despite several 2D and 3D structure prediction tools proposed in recent years, one of the challenges is to choose the best combination for accurate RNA 3D structure prediction. Here, we benchmarked seven small RNA PDB structures (40 to 90 nucleotides) with different topologies to understand the effects of different 2D structure predictions on the accuracy of 3D modeling. The current study explores the blind challenge of 2D to 3D conversions and highlights the performances of de novo RNA 3D modeling from their predicted 2D structure constraints. Our results show that conformational sampling-based methods such as SimRNA and IsRNA1 depend less on 2D accuracy, whereas motif-based methods account for 2D evidence. Our observations illustrate the disparities in available 3D and 2D prediction methods and may further offer insights into developing topology-specific or family-specific RNA structure prediction pipelines.
Copyright: © 2023 Kulkarni et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



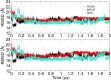
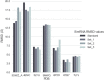
References
-
- Orland H, Zee A. RNA folding and large N matrix theory. Nucl Phys B. 2002;620: 456–476. 10.1016/S0550-3213(01)00522-3 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

