Mettl3-catalyzed m6A regulates histone modifier and modification expression in self-renewing somatic tissue
- PMID: 37656787
- PMCID: PMC10854438
- DOI: 10.1126/sciadv.adg5234
Mettl3-catalyzed m6A regulates histone modifier and modification expression in self-renewing somatic tissue
Abstract
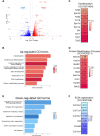
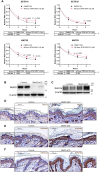
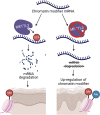
N6-methyladenosine (m6A) is the most abundant modification on messenger RNAs (mRNAs) and is catalyzed by methyltransferase-like protein 3 (Mettl3). To understand the role of m6A in a self-renewing somatic tissue, we deleted Mettl3 in epidermal progenitors in vivo. Mice lacking Mettl3 demonstrate marked features of dysfunctional development and self-renewal, including a loss of hair follicle morphogenesis and impaired cell adhesion and polarity associated with oral ulcerations. We show that Mettl3 promotes the m6A-mediated degradation of mRNAs encoding critical histone modifying enzymes. Depletion of Mettl3 results in the loss of m6A on these mRNAs and increases their expression and associated modifications, resulting in widespread gene expression abnormalities that mirror the gross phenotypic abnormalities. Collectively, these results have identified an additional layer of gene regulation within epithelial tissues, revealing an essential role for m6A in the regulation of chromatin modifiers, and underscoring a critical role for Mettl3-catalyzed m6A in proper epithelial development and self-renewal.
Figures


References
-
- A. Baroni, E. Buommino, V. de Gregorio, E. Ruocco, V. Ruocco, R. Wolf, Structure and function of the epidermis related to barrier properties. Clin. Dermatol. 30, 257–262 (2012). - PubMed
-
- P. Rousselle, E. Gentilhomme, Y. Neveux, Markers of epidermal proliferation and differentiation, in Agache’s Measuring the Skin, P. Humbert, F. Fanian, H. Maibach, P. Agache, Eds. (Springer International Publishing, 2017), pp. 397–405.
-
- E. M. Novoa, C. E. Mason, J. S. Mattick, Charting the unknown epitranscriptome. Nat. Rev. Mol. Cell Biol. 18, 339–340 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

