Proteomic and phosphoproteomic analyses of myectomy tissue reveals difference between sarcomeric and genotype-negative hypertrophic cardiomyopathy
- PMID: 37658118
- PMCID: PMC10474105
- DOI: 10.1038/s41598-023-40795-1
Proteomic and phosphoproteomic analyses of myectomy tissue reveals difference between sarcomeric and genotype-negative hypertrophic cardiomyopathy
Abstract
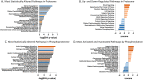
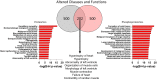
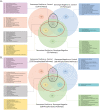
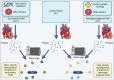
Hypertrophic cardiomyopathy (HCM) is a genetically heterogenous condition with about half of cases remaining genetically elusive or non-genetic in origin. HCM patients with a positive genetic test (HCMSarc) present earlier and with more severe disease than those with a negative genetic test (HCMNeg). We hypothesized these differences may be due to and/or reflect proteomic and phosphoproteomic differences between the two groups. TMT-labeled mass spectrometry was performed on 15 HCMSarc, 8 HCMNeg, and 7 control samples. There were 243 proteins differentially expressed and 257 proteins differentially phosphorylated between HCMSarc and HCMNeg. About 90% of pathways altered between genotypes were in disease-related pathways and HCMSarc showed enhanced proteomic and phosphoproteomic alterations in these pathways. Thus, we show HCMSarc has enhanced proteomic and phosphoproteomic dysregulation observed which may contribute to the more severe disease phenotype.
© 2023. Springer Nature Limited.
Conflict of interest statement
MJA is a consultant for Abbott, Boston Scientific, Bristol Myers Squibb, Daiichi Sankyo, Invitae, Medtronic, Tenaya Therapeutics, and UpToDate. MJA and Mayo Clinic have a royalty/equity relationship with AliveCor, Anumana, ARMGO Pharma, Pfizer, and Thryv Therapeutics. However, none of these entities have contributed to this study in any manner. The remaining authors have no conflicts to declare.
Figures




Similar articles
-
Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics.Proc Natl Acad Sci U S A. 2020 Oct 6;117(40):24691-24700. doi: 10.1073/pnas.2006764117. Epub 2020 Sep 23. Proc Natl Acad Sci U S A. 2020. PMID: 32968017 Free PMC article.
-
Multi-Omic Architecture of Obstructive Hypertrophic Cardiomyopathy.Circ Genom Precis Med. 2023 Apr;16(2):e003756. doi: 10.1161/CIRCGEN.122.003756. Epub 2023 Feb 20. Circ Genom Precis Med. 2023. PMID: 36802768
-
Influence of centre expertise on the diagnosis and management of hypertrophic cardiomyopathy: A study from the French register of hypertrophic cardiomyopathy (REMY).Int J Cardiol. 2019 Jan 15;275:107-113. doi: 10.1016/j.ijcard.2018.09.083. Epub 2018 Sep 28. Int J Cardiol. 2019. PMID: 30316646
-
Mass-Spectrometry-Based Functional Proteomic and Phosphoproteomic Technologies and Their Application for Analyzing Ex Vivo and In Vitro Models of Hypertrophic Cardiomyopathy.Int J Mol Sci. 2021 Dec 20;22(24):13644. doi: 10.3390/ijms222413644. Int J Mol Sci. 2021. PMID: 34948439 Free PMC article. Review.
-
Clinical outcomes associated with sarcomere mutations in hypertrophic cardiomyopathy: a meta-analysis on 7675 individuals.Clin Res Cardiol. 2018 Jan;107(1):30-41. doi: 10.1007/s00392-017-1155-5. Epub 2017 Aug 24. Clin Res Cardiol. 2018. PMID: 28840316 Review.
Cited by
-
Identification of candidate cardiomyopathy modifier genes through genome sequencing and RNA profiling.Front Cardiovasc Med. 2025 Jul 28;12:1546493. doi: 10.3389/fcvm.2025.1546493. eCollection 2025. Front Cardiovasc Med. 2025. PMID: 40791945 Free PMC article.
-
The molecular and cellular landscape of hypertrophic cardiomyopathy phenotypes: transition from obstructive to end-stage heart failure.J Mol Med (Berl). 2025 Jan;103(1):113-123. doi: 10.1007/s00109-024-02508-7. Epub 2025 Jan 7. J Mol Med (Berl). 2025. PMID: 39774683
-
Transcriptomic analysis and machine learning modeling identifies novel biomarkers and genetic characteristics of hypertrophic cardiomyopathy.Front Genet. 2025 Jun 17;16:1596049. doi: 10.3389/fgene.2025.1596049. eCollection 2025. Front Genet. 2025. PMID: 40599543 Free PMC article.
-
Proteomic Characterisation of Heart Failure Reveals a Unique Molecular Phenotype for Hypertrophic Cardiomyopathy.Biomedicines. 2024 Aug 1;12(8):1712. doi: 10.3390/biomedicines12081712. Biomedicines. 2024. PMID: 39200175 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

