Comparison of Four Protocols for In Vitro Differentiation of Human Embryonic Stem Cells into Trophoblast Lineages by BMP4 and Dual Inhibition of Activin/Nodal and FGF2 Signaling
- PMID: 37658178
- PMCID: PMC10784360
- DOI: 10.1007/s43032-023-01334-5
Comparison of Four Protocols for In Vitro Differentiation of Human Embryonic Stem Cells into Trophoblast Lineages by BMP4 and Dual Inhibition of Activin/Nodal and FGF2 Signaling
Abstract
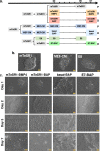
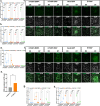
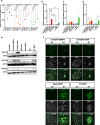
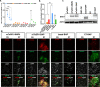
Human embryonic stem cells (hESCs) cultured in media containing bone morphogenic protein 4 (BMP4; B) differentiate into trophoblast-like cells. Supplementing media with inhibitors of activin/nodal signaling (A83-01) and of fibroblast growth factor 2 (PD173074) suppresses mesoderm and endoderm formation and improves specification of trophoblast-like lineages, but with variable effectiveness. We compared differentiation in four BMP4-containing media: mTeSR1-BMP4 only, mTeSR1-BAP, basal medium with BAP (basal-BAP), and a newly defined medium, E7-BAP. These media variably drive early differentiation towards trophoblast-like lineages with upregulation of early trophoblast markers CDX2 and KRT7 and downregulation of pluripotency markers (OCT4 and NANOG). As expected, based on differences between media in FGF2 and its inhibitors, downregulation of mesendoderm marker EOMES was variable between media. By day 7, only hESCs grown in E7-BAP or basal-BAP expressed HLA-G protein, indicating the presence of cells with extravillous trophoblast characteristics. Expression of HLA-G and other differentiation markers (hCG, KRT7, and GCM1) was highest in basal-BAP, suggesting a faster differentiation in this medium, but those cultures were more inhomogeneous and still expressed some endodermal and pluripotency markers. In E7-BAP, HLA-G expression increased later and was lower. There was also a low but maintained expression of some C19MC miRNAs, with more CpG hypomethylation of the ELF5 promoter, suggesting that E7-BAP cultures differentiate slower along the trophoblast lineage. We conclude that while all protocols drive differentiation into trophoblast lineages with varying efficiency, they have advantages and disadvantages that must be considered when selecting a protocol for specific experiments.
Keywords: Differentiation; Human embryonic stem cells; Syncytiotrophoblast, extravillous trophoblast; Trophoblast lineage.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Complete and unidirectional conversion of human embryonic stem cells to trophoblast by BMP4.Proc Natl Acad Sci U S A. 2013 Mar 26;110(13):E1212-21. doi: 10.1073/pnas.1303094110. Epub 2013 Mar 14. Proc Natl Acad Sci U S A. 2013. PMID: 23493551 Free PMC article.
-
Optimizing bone morphogenic protein 4-mediated human embryonic stem cell differentiation into trophoblast-like cells using fibroblast growth factor 2 and transforming growth factor-β/activin/nodal signalling inhibition.Reprod Biomed Online. 2017 Sep;35(3):253-263. doi: 10.1016/j.rbmo.2017.06.003. Epub 2017 Jun 12. Reprod Biomed Online. 2017. PMID: 28647356
-
Heightened potency of human pluripotent stem cell lines created by transient BMP4 exposure.Proc Natl Acad Sci U S A. 2015 May 5;112(18):E2337-46. doi: 10.1073/pnas.1504778112. Epub 2015 Apr 13. Proc Natl Acad Sci U S A. 2015. PMID: 25870291 Free PMC article.
-
Model systems for studying trophoblast differentiation from human pluripotent stem cells.Cell Tissue Res. 2012 Sep;349(3):809-24. doi: 10.1007/s00441-012-1371-2. Epub 2012 Mar 17. Cell Tissue Res. 2012. PMID: 22427062 Free PMC article. Review.
-
Trophoblast differentiation of human embryonic stem cells.Biotechnol J. 2013 Apr;8(4):421-33. doi: 10.1002/biot.201200203. Epub 2013 Jan 17. Biotechnol J. 2013. PMID: 23325630 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

