Recent advances in fungal xenobiotic metabolism: enzymes and applications
- PMID: 37658215
- PMCID: PMC10474215
- DOI: 10.1007/s11274-023-03737-7
Recent advances in fungal xenobiotic metabolism: enzymes and applications
Abstract
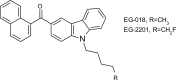
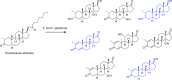
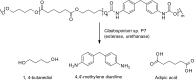
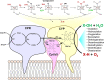
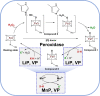
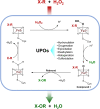
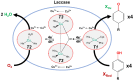
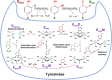
Fungi have been extensively studied for their capacity to biotransform a wide range of natural and xenobiotic compounds. This versatility is a reflection of the broad substrate specificity of fungal enzymes such as laccases, peroxidases and cytochromes P450, which are involved in these reactions. This review gives an account of recent advances in the understanding of fungal metabolism of drugs and pollutants such as dyes, agrochemicals and per- and poly-fluorinated alkyl substances (PFAS), and describes the key enzymes involved in xenobiotic biotransformation. The potential of fungi and their enzymes in the bioremediation of polluted environments and in the biocatalytic production of important compounds is also discussed.
Keywords: Biodegradation; Biotransformation; Lignin-degrading enzyme; Organic compounds; Pollutant.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Agrawal K, Chaturvedi V, Verma P. Fungal laccase discovered but yet undiscovered. Bioresour Bioprocess. 2018;5(1):4. doi: 10.1186/s40643-018-0190-z. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

