Intestinal microbiome and metabolome signatures in patients with chronic granulomatous disease
- PMID: 37659505
- PMCID: PMC11279821
- DOI: 10.1016/j.jaci.2023.07.022
Intestinal microbiome and metabolome signatures in patients with chronic granulomatous disease
Abstract
Background: Chronic granulomatous disease (CGD) is caused by defects in any 1 of the 6 subunits forming the nicotinamide adenine dinucleotide phosphate oxidase complex 2 (NOX2), leading to severely reduced or absent phagocyte-derived reactive oxygen species production. Almost 50% of patients with CGD have inflammatory bowel disease (CGD-IBD). While conventional IBD therapies can treat CGD-IBD, their benefits must be weighed against the risk of infection. Understanding the impact of NOX2 defects on the intestinal microbiota may lead to the identification of novel CGD-IBD treatments.
Objective: We sought to identify microbiome and metabolome signatures that can distinguish individuals with CGD and CGD-IBD.
Methods: We conducted a cross-sectional observational study of 79 patients with CGD, 8 pathogenic variant carriers, and 19 healthy controls followed at the National Institutes of Health Clinical Center. We profiled the intestinal microbiome (amplicon sequencing) and stool metabolome, and validated our findings in a second cohort of 36 patients with CGD recruited through the Primary Immune Deficiency Treatment Consortium.
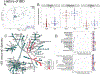
Results: We identified distinct intestinal microbiome and metabolome profiles in patients with CGD compared to healthy individuals. We observed enrichment for Erysipelatoclostridium spp, Sellimonas spp, and Lachnoclostridium spp in CGD stool samples. Despite differences in bacterial alpha and beta diversity between the 2 cohorts, several taxa correlated significantly between both cohorts. We further demonstrated that patients with CGD-IBD have a distinct microbiome and metabolome profile compared to patients without CGD-IBD.
Conclusion: Intestinal microbiome and metabolome signatures distinguished patients with CGD and CGD-IBD, and identified potential biomarkers and therapeutic targets.
Trial registration: ClinicalTrials.gov NCT02082353.
Keywords: CGD; Chronic granulomatous disease; IBD; NADPH oxidase; dysbiosis; inborn errors of immunity; inflammatory bowel disease; intestinal inflammation; metabolome; microbiome; primary immune deficiency.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

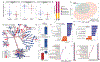




References
-
- Dhillon SS, Fattouh R, Elkadri A, Xu W, Murchie R, Walters T, et al. Variants in nicotinamide adenine dinucleotide phosphate oxidase complex components determine susceptibility to very early onset inflammatory bowel disease. Gastroenterology 2014; 147:680–9 e2. - PubMed
-
- Falcone EL, Holland SM. Gastrointestinal Complications in Chronic Granulomatous Disease. Methods Mol Biol 2019; 1982:573–86. - PubMed
-
- Magnani A, Brosselin P, Beaute J, de Vergnes N, Mouy R, Debre M, et al. Inflammatory manifestations in a single-center cohort of patients with chronic granulomatous disease. J Allergy Clin Immunol 2014; 134:655–62 e8. - PubMed
-
- Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000; 79:170–200. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

