Structure-based design of a strain transcending AMA1-RON2L malaria vaccine
- PMID: 37660103
- PMCID: PMC10475129
- DOI: 10.1038/s41467-023-40878-7
Structure-based design of a strain transcending AMA1-RON2L malaria vaccine
Abstract
Apical membrane antigen 1 (AMA1) is a key malaria vaccine candidate and target of neutralizing antibodies. AMA1 binds to a loop in rhoptry neck protein 2 (RON2L) to form the moving junction during parasite invasion of host cells, and this complex is conserved among apicomplexan parasites. AMA1-RON2L complex immunization achieves higher growth inhibitory activity than AMA1 alone and protects mice against Plasmodium yoelii challenge. Here, three single-component AMA1-RON2L immunogens were designed that retain the structure of the two-component AMA1-RON2L complex: one structure-based design (SBD1) and two insertion fusions. All immunogens elicited high antibody titers with potent growth inhibitory activity, yet these antibodies did not block RON2L binding to AMA1. The SBD1 immunogen induced significantly more potent strain-transcending neutralizing antibody responses against diverse strains of Plasmodium falciparum than AMA1 or AMA1-RON2L complex vaccination. This indicates that SBD1 directs neutralizing antibody responses to strain-transcending epitopes in AMA1 that are independent of RON2L binding. This work underscores the importance of neutralization mechanisms that are distinct from RON2 blockade. The stable single-component SBD1 immunogen elicits potent strain-transcending protection that may drive the development of next-generation vaccines for improved malaria and apicomplexan parasite control.
© 2023. Springer Nature Limited.
Conflict of interest statement
N.H.T., T.H.D., and P.N.P. are listed as inventors on a provisional patent application related to this work. The remaining authors declare no competing interests.
Figures
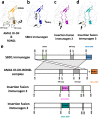
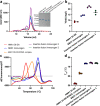

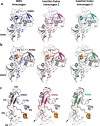
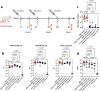
Similar articles
-
A strain-transcending anti-AMA1 human monoclonal antibody neutralizes malaria parasites independent of direct RON2L receptor blockade.Cell Rep Med. 2025 Mar 18;6(3):101985. doi: 10.1016/j.xcrm.2025.101985. Epub 2025 Feb 27. Cell Rep Med. 2025. PMID: 40020675 Free PMC article.
-
Structure guided mimicry of an essential P. falciparum receptor-ligand complex enhances cross neutralizing antibodies.Nat Commun. 2023 Sep 21;14(1):5879. doi: 10.1038/s41467-023-41636-5. Nat Commun. 2023. PMID: 37735574 Free PMC article.
-
A malaria vaccine protects Aotus monkeys against virulent Plasmodium falciparum infection.NPJ Vaccines. 2017;2:14. doi: 10.1038/s41541-017-0015-7. Epub 2017 May 22. NPJ Vaccines. 2017. PMID: 28804644 Free PMC article.
-
Apical membrane antigen 1 as an anti-malarial drug target.Curr Top Med Chem. 2011;11(16):2039-47. doi: 10.2174/156802611796575885. Curr Top Med Chem. 2011. PMID: 21619512 Review.
-
Apical membrane antigen 1: a malaria vaccine candidate in review.Trends Parasitol. 2008 Feb;24(2):74-84. doi: 10.1016/j.pt.2007.12.002. Epub 2008 Jan 15. Trends Parasitol. 2008. PMID: 18226584 Review.
Cited by
-
Malaria vaccines: a new era of prevention and control.Nat Rev Microbiol. 2024 Dec;22(12):756-772. doi: 10.1038/s41579-024-01065-7. Epub 2024 Jul 18. Nat Rev Microbiol. 2024. PMID: 39025972 Review.
-
The Need for Novel Asexual Blood-Stage Malaria Vaccine Candidates for Plasmodium falciparum.Biomolecules. 2024 Jan 12;14(1):100. doi: 10.3390/biom14010100. Biomolecules. 2024. PMID: 38254700 Free PMC article. Review.
-
Holding hands to halt malaria: stronger together through heterotypic antibody interactions.Trends Parasitol. 2025 May;41(5):337-338. doi: 10.1016/j.pt.2025.04.004. Epub 2025 Apr 22. Trends Parasitol. 2025. PMID: 40268600
-
Potent AMA1-specific human monoclonal antibody against Plasmodium vivax Pre-erythrocytic and Blood Stages.Nat Commun. 2024 Dec 4;15(1):10556. doi: 10.1038/s41467-024-53848-4. Nat Commun. 2024. PMID: 39632799 Free PMC article.
-
Search continues: Exploring immunoinformatics platforms for designing an effective multiepitope malaria vaccine candidate.BioTechnologia (Pozn). 2025 Jun 30;106(2):151-168. doi: 10.5114/bta/204528. eCollection 2025. BioTechnologia (Pozn). 2025. PMID: 40703713 Free PMC article.
References
-
- World Health Organisation. World Malaria Report (WHO, Geneva, 2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

