Ufmylation on UFBP1 alleviates non-alcoholic fatty liver disease by modulating hepatic endoplasmic reticulum stress
- PMID: 37660122
- PMCID: PMC10475044
- DOI: 10.1038/s41419-023-06095-2
Ufmylation on UFBP1 alleviates non-alcoholic fatty liver disease by modulating hepatic endoplasmic reticulum stress
Abstract
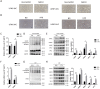
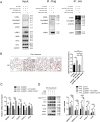
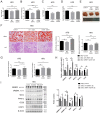
Non-alcoholic fatty liver disease (NAFLD) is the most common liver disease characterized by lipid accumulation and endoplasmic reticulum (ER) stress, while effective therapies targeting the specific characteristics of NAFLD are limited. Ufmylation is a newly found post-translational modification process that involves the attachment of the Ubiquitin-fold modifier 1 (UFM1) protein to its substrates via ufmylation modification system. Ufmylation regulates ER stress via modifying UFM1 binding protein 1 (UFBP1), suggesting a potential role for ufmylation in NAFLD pathogenesis. However, the precise role of ufmylation in NAFLD remains unclear. Herein, we aim to elucidate the impact of ufmylation on UFBP1 in NAFLD and explore the underlying mechanisms involved. We observed increased expression of UFM1-conjugated proteins and ufmylation modification system components in livers with steatosis derived from NAFLD patients and NAFLD models. Upregulation of ufmylation on hepatic proteins appeared to be an adaptive response to hepatic ER stress in NAFLD. In vitro, knocking down UFBP1 resulted in increased lipid accumulation and lipogenesis in hepatocytes treated with free fatty acids (FFA), which could be rescued by wild-type UFBP1 (WT UFBP1) but not by a mutant form of UFBP1 lacking the main ufmylation site lys267 (UFBP1 K267R). In vivo, ufmylation on UFBP1 ameliorated obesity, hepatic steatosis, hepatic lipogenesis, dyslipidemia, insulin resistance and liver damage in mice with NAFLD induced by a high fat diet (HFD). We also demonstrated that the downregulation of UFBP1 induced ER stress, whereas the reintroduction or overexpression of UFBP1 alleviated ER stress in a manner dependent on ufmylation in NAFLD. This mechanism could be responsible for the amelioration of aberrant hepatic lipogenesis and insulin resistance in NAFLD. Our data reveal a protective role of ufmylation on UFBP1 against NAFLD and offer a specific target for NAFLD treatment.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

