Theranostic imaging and multimodal photodynamic therapy and immunotherapy using the mTOR signaling pathway
- PMID: 37660174
- PMCID: PMC10475087
- DOI: 10.1038/s41467-023-40826-5
Theranostic imaging and multimodal photodynamic therapy and immunotherapy using the mTOR signaling pathway
Abstract
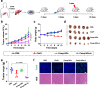
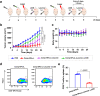
Tumor metastases are considered the leading cause of cancer-associated deaths. While clinically applied drugs have demonstrated to efficiently remove the primary tumor, metastases remain poorly accessible. To overcome this limitation, herein, the development of a theranostic nanomaterial by incorporating a chromophore for imaging and a photosensitizer for treatment of metastatic tumor sites is presented. The mechanism of action reveals that the nanoparticles are able to intervene by local generation of cellular damage through photodynamic therapy as well as by systemic induction of an immune response by immunotherapy upon inhibition of the mTOR signaling pathway which is of crucial importance for tumor onset, progression and metastatic spreading. The nanomaterial is able to strongly reduce the volume of the primary tumor as well as eradicates tumor metastases in a metastatic breast cancer and a multi-drug resistant patient-derived hepatocellular carcinoma models in female mice.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. - PubMed
-
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. - PubMed
-
- Nishikawa M. Reactive oxygen species in tumor metastasis. Cancer Lett. 2008;266:53–59. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

