The RavA-ViaA chaperone complex modulates bacterial persistence through its association with the fumarate reductase enzyme
- PMID: 37660904
- PMCID: PMC10585395
- DOI: 10.1016/j.jbc.2023.105199
The RavA-ViaA chaperone complex modulates bacterial persistence through its association with the fumarate reductase enzyme
Abstract
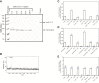
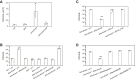
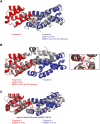
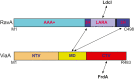
Regulatory ATPase variant A (RavA) is a MoxR AAA+ protein that functions together with a partner protein termed von Willebrand factor type A interacting with AAA+ ATPase (ViaA). RavA-ViaA are functionally associated with anaerobic respiration in Escherichia coli through interactions with the fumarate reductase (Frd) electron transport complex. Through this association, RavA and ViaA modulate the activity of the Frd complex and, hence, are proposed to have chaperone-like activity. However, the functional role of RavA-ViaA in the cell is not yet well established. We had demonstrated that RavA-ViaA can sensitize E. coli cells to sublethal concentrations of the aminoglycoside class of antibiotics. Since Frd has been associated with bacterial persistence against antibiotics, the relationship of RavA-ViaA and Frd was explored within this context. Experiments performed here reveal a function of RavA-ViaA in bacterial persistence upon treatment with antibiotics through the association of the chaperone complex with Frd. As part of this work, the NMR structure of the N-terminal domain of ViaA was solved. The structure reveals a novel alpha helical fold, which we name the VAN fold, that has not been observed before. We show that this domain is required for the function of the chaperone complex. We propose that modulating the levels of RavA-ViaA could enhance the susceptibility of Gram-negative bacteria to antibiotics.
Keywords: AAA+ proteins; MoxR; NMR structure; RavA; ViaA; bacterial persistence; von Willebrand factor type A domain.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Similar articles
-
The RavA-ViaA Chaperone-Like System Interacts with and Modulates the Activity of the Fumarate Reductase Respiratory Complex.J Mol Biol. 2017 Jan 20;429(2):324-344. doi: 10.1016/j.jmb.2016.12.008. Epub 2016 Dec 12. J Mol Biol. 2017. PMID: 27979649
-
Bioenergetic State of Escherichia coli Controls Aminoglycoside Susceptibility.mBio. 2023 Feb 28;14(1):e0330222. doi: 10.1128/mbio.03302-22. Epub 2023 Jan 10. mBio. 2023. PMID: 36625597 Free PMC article.
-
The MoxR ATPase RavA and its cofactor ViaA interact with the NADH:ubiquinone oxidoreductase I in Escherichia coli.PLoS One. 2014 Jan 15;9(1):e85529. doi: 10.1371/journal.pone.0085529. eCollection 2014. PLoS One. 2014. PMID: 24454883 Free PMC article.
-
Novel structural and functional insights into the MoxR family of AAA+ ATPases.J Struct Biol. 2012 Aug;179(2):211-21. doi: 10.1016/j.jsb.2012.03.010. Epub 2012 Apr 3. J Struct Biol. 2012. PMID: 22491058 Review.
-
Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation.Crit Rev Biochem Mol Biol. 2007 Mar-Apr;42(2):95-111. doi: 10.1080/10409230701322298. Crit Rev Biochem Mol Biol. 2007. PMID: 17453917 Review.
Cited by
-
Molecular Characterization of the MoxR AAA+ ATPase of Synechococcus sp. Strain NKBG15041c.Int J Mol Sci. 2024 Sep 15;25(18):9955. doi: 10.3390/ijms25189955. Int J Mol Sci. 2024. PMID: 39337443 Free PMC article.
References
-
- Bhandari V., Van Ommen D.A.J., Wong K.S., Houry W.A. Analysis of the evolution of the MoxR ATPases. J. Phys. Chem. A. 2022;126:4734–4746. - PubMed
-
- Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. - PubMed
-
- Kahle M., Ter Beek J., Hosler J.P., Adelroth P. The insertion of the non-heme Fe(B) cofactor into nitric oxide reductase from P. denitrificans depends on NorQ and NorD accessory proteins. Biochim. Biophys. Acta Bioenerg. 2018;1859:1051–1058. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

