Structure and function analysis of a type III preQ1-I riboswitch from Escherichia coli reveals direct metabolite sensing by the Shine-Dalgarno sequence
- PMID: 37660906
- PMCID: PMC10622847
- DOI: 10.1016/j.jbc.2023.105208
Structure and function analysis of a type III preQ1-I riboswitch from Escherichia coli reveals direct metabolite sensing by the Shine-Dalgarno sequence
Abstract
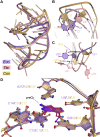
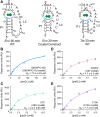
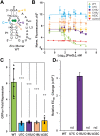
Riboswitches are small noncoding RNAs found primarily in the 5' leader regions of bacterial messenger RNAs where they regulate expression of downstream genes in response to binding one or more cellular metabolites. Such noncoding RNAs are often regulated at the translation level, which is thought to be mediated by the accessibility of the Shine-Dalgarno sequence (SDS) ribosome-binding site. Three classes (I-III) of prequeuosine1 (preQ1)-sensing riboswitches are known that control translation. Class I is divided into three subtypes (types I-III) that have diverse mechanisms of sensing preQ1, which is involved in queuosine biosynthesis. To provide insight into translation control, we determined a 2.30 Å-resolution cocrystal structure of a class I type III preQ1-sensing riboswitch identified in Escherichia coli (Eco) by bioinformatic searches. The Eco riboswitch structure differs from previous preQ1 riboswitch structures because it has the smallest naturally occurring aptamer and the SDS directly contacts the preQ1 metabolite. We validated structural observations using surface plasmon resonance and in vivo gene-expression assays, which showed strong switching in live E. coli. Our results demonstrate that the Eco riboswitch is relatively sensitive to mutations that disrupt noncanonical interactions that form the pseudoknot. In contrast to type II preQ1 riboswitches, a kinetic analysis showed that the type III Eco riboswitch strongly prefers preQ1 over the chemically similar metabolic precursor preQ0. Our results reveal the importance of noncanonical interactions in riboswitch-driven gene regulation and the versatility of the class I preQ1 riboswitch pseudoknot as a metabolite-sensing platform that supports SDS sequestration.
Keywords: Shine-Dalgarno sequence; gene regulation; molecular recognition; preQ(1)-I riboswitch; pseudoknot; translation regulation; x-ray crystallography.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures





Similar articles
-
Knotty is nice: Metabolite binding and RNA-mediated gene regulation by the preQ1 riboswitch family.J Biol Chem. 2024 Dec;300(12):107951. doi: 10.1016/j.jbc.2024.107951. Epub 2024 Oct 30. J Biol Chem. 2024. PMID: 39486689 Free PMC article. Review.
-
Nucleobase mutants of a bacterial preQ1-II riboswitch that uncouple metabolite sensing from gene regulation.J Biol Chem. 2020 Feb 28;295(9):2555-2567. doi: 10.1074/jbc.RA119.010755. Epub 2019 Oct 28. J Biol Chem. 2020. PMID: 31659117 Free PMC article.
-
Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics.Proc Natl Acad Sci U S A. 2015 Jul 7;112(27):E3485-94. doi: 10.1073/pnas.1503955112. Epub 2015 Jun 23. Proc Natl Acad Sci U S A. 2015. PMID: 26106162 Free PMC article.
-
Structural determinants for ligand capture by a class II preQ1 riboswitch.Proc Natl Acad Sci U S A. 2014 Feb 11;111(6):E663-71. doi: 10.1073/pnas.1400126111. Epub 2014 Jan 27. Proc Natl Acad Sci U S A. 2014. PMID: 24469808 Free PMC article.
-
Structure and function of preQ1 riboswitches.Biochim Biophys Acta. 2014 Oct;1839(10):939-950. doi: 10.1016/j.bbagrm.2014.04.019. Epub 2014 May 4. Biochim Biophys Acta. 2014. PMID: 24798077 Free PMC article. Review.
Cited by
-
fpocketR: A platform for identification and analysis of ligand-binding pockets in RNA.bioRxiv [Preprint]. 2025 Mar 29:2025.03.25.645323. doi: 10.1101/2025.03.25.645323. bioRxiv. 2025. PMID: 40196532 Free PMC article. Preprint.
-
Engineering covalent small molecule-RNA complexes in living cells.Nat Chem Biol. 2025 Jun;21(6):843-854. doi: 10.1038/s41589-024-01801-3. Epub 2025 Jan 6. Nat Chem Biol. 2025. PMID: 39762536 Free PMC article.
-
Comparative analysis of RNA 3D structure prediction methods: towards enhanced modeling of RNA-ligand interactions.Nucleic Acids Res. 2024 Jul 22;52(13):7465-7486. doi: 10.1093/nar/gkae541. Nucleic Acids Res. 2024. PMID: 38917327 Free PMC article.
-
Two riboswitch classes that share a common ligand-binding fold show major differences in the ability to accommodate mutations.Nucleic Acids Res. 2024 Nov 27;52(21):13152-13173. doi: 10.1093/nar/gkae886. Nucleic Acids Res. 2024. PMID: 39413212 Free PMC article.
-
Knotty is nice: Metabolite binding and RNA-mediated gene regulation by the preQ1 riboswitch family.J Biol Chem. 2024 Dec;300(12):107951. doi: 10.1016/j.jbc.2024.107951. Epub 2024 Oct 30. J Biol Chem. 2024. PMID: 39486689 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

