Transcription factor UBF depletion in mouse cells results in downregulation of both downstream and upstream elements of the rRNA transcription network
- PMID: 37660911
- PMCID: PMC10558777
- DOI: 10.1016/j.jbc.2023.105203
Transcription factor UBF depletion in mouse cells results in downregulation of both downstream and upstream elements of the rRNA transcription network
Abstract
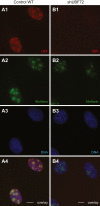
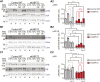
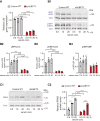
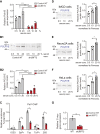
Transcription/processing of the ribosomal RNA (rRNA) precursor, as part of ribosome biosynthesis, is intensively studied and characterized in eukaryotic cells. Here, we constructed shRNA-based mouse cell lines partially silenced for the Upstream Binding Factor UBF, the master regulator of rRNA transcription and organizer of open rDNA chromatin. Full Ubf silencing in vivo is not viable, and these new tools allow further characterization of rRNA transcription and its coordination with cellular signaling. shUBF cells display cell cycle G1 delay and reduced 47S rRNA precursor and 28S rRNA at baseline and serum-challenged conditions. Growth-related mTOR signaling is downregulated with the fractions of active phospho-S6 Kinase and pEIF4E translation initiation factor reduced, similar to phosphorylated cell cycle regulator retinoblastoma, pRB, positive regulator of UBF availability/rRNA transcription. Additionally, we find transcription-competent pUBF (Ser484) severely restricted and its interacting initiation factor RRN3 reduced and responsive to extracellular cues. Furthermore, fractional UBF occupancy on the rDNA unit is decreased in shUBF, and expression of major factors involved in different aspects of rRNA transcription is severely downregulated by UBF depletion. Finally, we observe reduced RNA Pol1 occupancy over rDNA promoter sequences and identified unexpected regulation of RNA Pol1 expression, relative to serum availability and under UBF silencing, suggesting that regulation of rRNA transcription may not be restricted to modulation of Pol1 promoter binding/elongation rate. Overall, this work reveals that UBF depletion has a critical downstream and upstream impact on the whole network orchestrating rRNA transcription in mammalian cells.
Keywords: C-MYC; RNA pol1; cellular growth; rRNA; transcriptional regulation.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

