Epizootiological surveillance of porcine circoviruses in free-ranging wild boars in China
- PMID: 37660950
- PMCID: PMC10590700
- DOI: 10.1016/j.virs.2023.08.008
Epizootiological surveillance of porcine circoviruses in free-ranging wild boars in China
Abstract
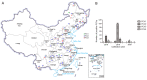
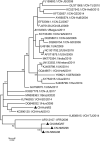
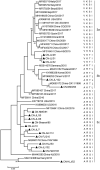
Four species of porcine circoviruses (PCV1-4) have been reported to circulate in Chinese domestic pigs, while the epizootiology of these viruses in free-ranging wild boars in China remains unknown. In this study, tissue and serum samples collected from diseased or apparently healthy wild boars between 2018 and 2020 in 19 regions of China were tested for the prevalence of PCV1-4 infections. Positive rates of PCV1, PCV2, and PCV3 DNA in the tissue samples of Chinese wild boars were 1.6% (4/247), 58.3% (144/247), and 10.9% (27/247) respectively, with none positive for PCV4. Sequence analysis of viral genome showed that the four PCV1 strains distributed in Hunan and Inner Mongolia shared 97.5%-99.6% sequence identity with global distributed reference strains. Comparison of the ORF2 gene sequences showed that 80 PCV2 strains widely distributed in 18 regions shared 79.5%-100% sequence identity with reference strains from domestic pigs and wild boars, and were grouped into PCV2a (7), PCV2b (31) and PCV2d (42). For PCV3, 17 sequenced strains shared 97.2%-100% nucleotide identity at the genomic level and could be divided into PCV3a (3), PCV3b (2) and PCV3c (12) based on the phylogeny of ORF2 gene sequences. Serological data revealed antibody positive rates against PCV1 and PCV2 of 11.4% (19/167) and 53.9% (90/167) respectively. The data obtained in this study improved our understanding about the epidemiological situations of PCVs infection in free-ranging wild boars in China and will be valuable for the prevention and control of diseases caused by PCVs infection.
Keywords: China; Epizootiology; Porcine circoviruses (PCVs); Wild boar.
Copyright © 2023 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Molecular epidemiological surveillance and complete genome analysis of porcine circoviruses in wild boars (Sus scrofa) in Gyeongnam Province, South Korea.J Vet Sci. 2024 Nov;25(6):e79. doi: 10.4142/jvs.24252. J Vet Sci. 2024. PMID: 39608773 Free PMC article.
-
First complete genomic sequence analysis of porcine circovirus type 4 (PCV4) in wild boars.Vet Microbiol. 2022 Oct;273:109547. doi: 10.1016/j.vetmic.2022.109547. Epub 2022 Aug 17. Vet Microbiol. 2022. PMID: 36037620
-
Comprehensive Survey of PCV2 and PCV3 in Domestic Pigs and Wild Boars Across Portugal: Prevalence, Geographical Distribution and Genetic Diversity.Pathogens. 2025 Jul 9;14(7):675. doi: 10.3390/pathogens14070675. Pathogens. 2025. PMID: 40732721 Free PMC article.
-
Porcine circoviruses: current status, knowledge gaps and challenges.Virus Res. 2020 Sep;286:198044. doi: 10.1016/j.virusres.2020.198044. Epub 2020 Jun 2. Virus Res. 2020. PMID: 32502553 Review.
-
Advances in Crosstalk between Porcine Circoviruses and Host.Viruses. 2022 Jun 28;14(7):1419. doi: 10.3390/v14071419. Viruses. 2022. PMID: 35891399 Free PMC article. Review.
Cited by
-
Genetic diversity of porcine circoviruses 2 and 3 circulating among wild boars in the Moscow Region of Russia.Front Vet Sci. 2024 Jun 26;11:1372203. doi: 10.3389/fvets.2024.1372203. eCollection 2024. Front Vet Sci. 2024. PMID: 38988985 Free PMC article.
-
Geographical distribution and characterization of Jingmen tick virus in wild boars in China.Virol Sin. 2025 Feb;40(1):137-140. doi: 10.1016/j.virs.2024.12.011. Epub 2025 Jan 1. Virol Sin. 2025. PMID: 39753193 Free PMC article.
-
Molecular epidemiological surveillance and complete genome analysis of porcine circoviruses in wild boars (Sus scrofa) in Gyeongnam Province, South Korea.J Vet Sci. 2024 Nov;25(6):e79. doi: 10.4142/jvs.24252. J Vet Sci. 2024. PMID: 39608773 Free PMC article.
-
The single-cell transcriptional landscape of lung cells from PCV2d-infected mice.Front Microbiol. 2025 Mar 24;16:1554961. doi: 10.3389/fmicb.2025.1554961. eCollection 2025. Front Microbiol. 2025. PMID: 40196036 Free PMC article.
-
Both recombinant Bacillus subtilis Expressing PCV2d Cap protein and PCV2d-VLPs can stimulate strong protective immune responses in mice.Heliyon. 2023 Nov 28;9(12):e22941. doi: 10.1016/j.heliyon.2023.e22941. eCollection 2023 Dec. Heliyon. 2023. PMID: 38058449 Free PMC article.
References
-
- Allan G., McNeilly F., Kennedy S., Daft B., Clarke E., Ellis J., Haines D., Meehan B., Adair B. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and europe. J. Vet. Diagn. Invest. 1998;10:3–10. - PubMed
-
- Amoroso M., Serra F., Esposito C., D'Alessio N., Ferrara G., Cioffi B., Anzalone A., Pagnini U., De Carlo E., Fusco G., Montagnaro S. Prevalence of infection with porcine circovirus types 2 and 3 in the wild boar population in the campania region (southern Italy) Animals. 2021;11:3215. - PMC - PubMed
-
- Barbosa C., Martins N., Freitas T., Lobato Z. Serological survey of porcine circovirus-2 in captive wild boars (sus scrofa) from registered farms of south and south-east regions of Brazil. Transbound. Emerg. Dis. 2016;63:e278–e280. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

