Whole genome sequence and diversity in multigene families of Babesia ovis
- PMID: 37662008
- PMCID: PMC10471129
- DOI: 10.3389/fcimb.2023.1194608
Whole genome sequence and diversity in multigene families of Babesia ovis
Abstract
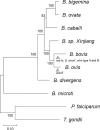
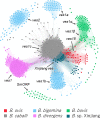
Ovine babesiosis, caused by Babesia ovis, is an acute, lethal, and endemic disease worldwide and causes a huge economic loss to animal industry. Pathogen genome sequences can be utilized for selecting diagnostic markers, drug targets, and antigens for vaccine development; however, those for B. ovis have not been available so far. In this study, we obtained a draft genome sequence for B. ovis isolated from an infected sheep in Turkey. The genome size was 7.81 Mbp with 3,419 protein-coding genes. It consisted of 41 contigs, and the N50 was 526 Kbp. There were 259 orthologs identified among eight Babesia spp., Plasmodium falciparum, and Toxoplasma gondii. A phylogeny was estimated on the basis of the orthologs, which showed B. ovis to be closest to B. bovis. There were 43 ves genes identified using hmm model as well. They formed a discriminating cluster to other ves multigene family of Babesia spp. but showed certain similarities to those of B. bovis, B. caballi, and Babesia sp. Xinjiang, which is consistent with the phylogeny. Comparative genomics among B. ovis and B. bovis elucidated uniquely evolved genes in these species, which may account for the adaptation.
Keywords: Babesia ovis; apicomplexa; comparative genomics; multigene families; ovine babesiosis.
Copyright © 2023 Yamagishi, Ceylan, Xuan and Sevinc.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Chromosome-level genome assembly of Babesia caballi reveals diversity of multigene families among Babesia species.BMC Genomics. 2023 Aug 24;24(1):483. doi: 10.1186/s12864-023-09540-w. BMC Genomics. 2023. PMID: 37620766 Free PMC article.
-
High genetic diversity and differentiation of the Babesia ovis population in Turkey.Transbound Emerg Dis. 2020 Jul;67 Suppl 2:26-35. doi: 10.1111/tbed.13174. Epub 2019 Jun 24. Transbound Emerg Dis. 2020. PMID: 31231917
-
Molecular detection and genetic characterization of Babesia, Theileria and Anaplasma amongst apparently healthy sheep and goats in the central region of Turkey.Ticks Tick Borne Dis. 2017 Feb;8(2):246-252. doi: 10.1016/j.ttbdis.2016.11.006. Epub 2016 Nov 16. Ticks Tick Borne Dis. 2017. PMID: 27908771
-
Comparative Bioinformatics Analysis of Transcription Factor Genes Indicates Conservation of Key Regulatory Domains among Babesia bovis, Babesia microti, and Theileria equi.PLoS Negl Trop Dis. 2016 Nov 10;10(11):e0004983. doi: 10.1371/journal.pntd.0004983. eCollection 2016 Nov. PLoS Negl Trop Dis. 2016. PMID: 27832060 Free PMC article. Review.
-
Genetic basis for GPI-anchor merozoite surface antigen polymorphism of Babesia and resulting antigenic diversity.Vet Parasitol. 2006 May 31;138(1-2):33-49. doi: 10.1016/j.vetpar.2006.01.038. Epub 2006 Mar 23. Vet Parasitol. 2006. PMID: 16551492 Review.
Cited by
-
ves1α genes expression is the major determinant of Babesia bovis-infected erythrocytes cytoadhesion to endothelial cells.PLoS Pathog. 2025 Apr 28;21(4):e1012583. doi: 10.1371/journal.ppat.1012583. eCollection 2025 Apr. PLoS Pathog. 2025. PMID: 40294074 Free PMC article.
References
-
- Altenhoff A. M., Škunca N., Glover N., Train C.-M., Sueki A., Piližota I., et al. . (2015). The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 43 (Database issue), D240–D249. doi: 10.1093/nar/gku1158 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

