Comparative genomics and DNA methylation analysis of Pseudomonas aeruginosa clinical isolate PA3 by single-molecule real-time sequencing reveals new targets for antimicrobials
- PMID: 37662009
- PMCID: PMC10471985
- DOI: 10.3389/fcimb.2023.1180194
Comparative genomics and DNA methylation analysis of Pseudomonas aeruginosa clinical isolate PA3 by single-molecule real-time sequencing reveals new targets for antimicrobials
Abstract
Introduction: Pseudomonas aeruginosa (P.aeruginosa) is an important opportunistic pathogen with broad environmental adaptability and complex drug resistance. Single-molecule real-time (SMRT) sequencing technique has longer read-length sequences, more accuracy, and the ability to identify epigenetic DNA alterations.
Methods: This study applied SMRT technology to sequence a clinical strain P. aeruginosa PA3 to obtain its genome sequence and methylation modification information. Genomic, comparative, pan-genomic, and epigenetic analyses of PA3 were conducted.
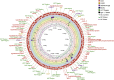
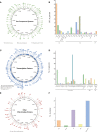
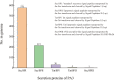
Results: General genome annotations of PA3 were discovered, as well as information about virulence factors, regulatory proteins (RPs), secreted proteins, type II toxin-antitoxin (TA) pairs, and genomic islands. A genome-wide comparison revealed that PA3 was comparable to other P. aeruginosa strains in terms of identity, but varied in areas of horizontal gene transfer (HGT). Phylogenetic analysis showed that PA3 was closely related to P. aeruginosa 60503 and P. aeruginosa 8380. P. aeruginosa's pan-genome consists of a core genome of roughly 4,300 genes and an accessory genome of at least 5,500 genes. The results of the epigenetic analysis identified one main methylation sites, N6-methyladenosine (m6A) and 1 motif (CATNNNNNNNTCCT/AGGANNNNNNNATG). 16 meaningful methylated sites were picked. Among these, purH, phaZ, and lexA are of great significance playing an important role in the drug resistance and biological environment adaptability of PA3, and the targeting of these genes may benefit further antibacterial studies.
Disucssion: This study provided a detailed visualization and DNA methylation information of the PA3 genome and set a foundation for subsequent research into the molecular mechanism of DNA methyltransferase-controlled P. aeruginosa pathogenicity.
Keywords: DNA methylation analysis; Pseudomonas aeruginosa; SMRT; antibacterial; comparative genome analysis; epigenetics.
Copyright © 2023 Li, Zhou, Liao, Liu, Zhao, Wang, Zhong, Zeng, Peng, Tan and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Genomic analyses of multidrug resistant Pseudomonas aeruginosa PA1 resequenced by single-molecule real-time sequencing.Biosci Rep. 2016 Nov 29;36(6):e00418. doi: 10.1042/BSR20160282. Print 2016 Dec. Biosci Rep. 2016. PMID: 27765811 Free PMC article.
-
DNA Methyltransferase Regulates Nitric Oxide Homeostasis and Virulence in a Chronically Adapted Pseudomonas aeruginosa Strain.mSystems. 2022 Oct 26;7(5):e0043422. doi: 10.1128/msystems.00434-22. Epub 2022 Sep 15. mSystems. 2022. PMID: 36106744 Free PMC article.
-
Identification of a Pseudomonas aeruginosa PAO1 DNA Methyltransferase, Its Targets, and Physiological Roles.mBio. 2017 Feb 21;8(1):e02312-16. doi: 10.1128/mBio.02312-16. mBio. 2017. PMID: 28223461 Free PMC article.
-
Genomic and Metabolic Characteristics of the Pathogenicity in Pseudomonas aeruginosa.Int J Mol Sci. 2021 Nov 29;22(23):12892. doi: 10.3390/ijms222312892. Int J Mol Sci. 2021. PMID: 34884697 Free PMC article. Review.
-
Carbapenem-Resistant Pseudomonas aeruginosa's Resistome: Pan-Genomic Plasticity, the Impact of Transposable Elements and Jumping Genes.Antibiotics (Basel). 2025 Mar 31;14(4):353. doi: 10.3390/antibiotics14040353. Antibiotics (Basel). 2025. PMID: 40298491 Free PMC article. Review.
Cited by
-
DNA methylome regulates virulence and metabolism in Pseudomonas syringae.Elife. 2025 Feb 24;13:RP96290. doi: 10.7554/eLife.96290. Elife. 2025. PMID: 39992965 Free PMC article.
-
Antibacterial Activity of Plants in Cirsium: A Comprehensive Review.Chin J Integr Med. 2024 Sep;30(9):835-841. doi: 10.1007/s11655-024-3757-2. Epub 2024 Mar 27. Chin J Integr Med. 2024. PMID: 38532154 Review.
-
Correlation Analysis Between Multi-Drug Resistance Phenotype and Virulence Factor Expression of Clinical Pseudomonas aeruginosa.Curr Issues Mol Biol. 2025 Jan 15;47(1):50. doi: 10.3390/cimb47010050. Curr Issues Mol Biol. 2025. PMID: 39852165 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

