This is a preprint.
Heterogeneous SARS-CoV-2 kinetics due to variable timing and intensity of immune responses
- PMID: 37662228
- PMCID: PMC10473815
- DOI: 10.1101/2023.08.20.23294350
Heterogeneous SARS-CoV-2 kinetics due to variable timing and intensity of immune responses
Update in
-
Heterogeneous SARS-CoV-2 kinetics due to variable timing and intensity of immune responses.JCI Insight. 2024 Apr 4;9(9):e176286. doi: 10.1172/jci.insight.176286. JCI Insight. 2024. PMID: 38573774 Free PMC article.
Abstract
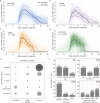
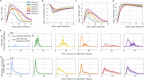
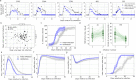
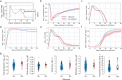
The viral kinetics of documented SARS-CoV-2 infections exhibit a high degree of inter-individual variability. We identified six distinct viral shedding patterns, which differed according to peak viral load, duration, expansion rate and clearance rate, by clustering data from 768 infections in the National Basketball Association cohort. Omicron variant infections in previously vaccinated individuals generally led to lower cumulative shedding levels of SARS-CoV-2 than other scenarios. We then developed a mechanistic mathematical model that recapitulated 1510 observed viral trajectories, including viral rebound and cases of reinfection. Lower peak viral loads were explained by a more rapid and sustained transition of susceptible cells to a refractory state during infection, as well as an earlier and more potent late, cytolytic immune response. Our results suggest that viral elimination occurs more rapidly during omicron infection, following vaccination, and following re-infection due to enhanced innate and acquired immune responses. Because viral load has been linked with COVID-19 severity and transmission risk, our model provides a framework for understanding the wide range of observed SARS-CoV-2 infection outcomes.
Figures


Similar articles
-
Heterogeneous SARS-CoV-2 kinetics due to variable timing and intensity of immune responses.JCI Insight. 2024 Apr 4;9(9):e176286. doi: 10.1172/jci.insight.176286. JCI Insight. 2024. PMID: 38573774 Free PMC article.
-
Viral kinetics of SARS-CoV-2 following onset of COVID-19 in symptomatic patients infected with the ancestral strain and omicron BA.2 in Hong Kong: a retrospective observational study.Lancet Microbe. 2023 Sep;4(9):e722-e731. doi: 10.1016/S2666-5247(23)00146-5. Lancet Microbe. 2023. PMID: 37659420
-
Viral and antibody dynamics of acute infection with SARS-CoV-2 omicron variant (B.1.1.529): a prospective cohort study from Shenzhen, China.Lancet Microbe. 2023 Aug;4(8):e632-e641. doi: 10.1016/S2666-5247(23)00139-8. Epub 2023 Jul 14. Lancet Microbe. 2023. PMID: 37459867 Review.
-
Transmission and Infectious SARS-CoV-2 Shedding Kinetics in Vaccinated and Unvaccinated Individuals.JAMA Netw Open. 2022 May 2;5(5):e2213606. doi: 10.1001/jamanetworkopen.2022.13606. JAMA Netw Open. 2022. PMID: 35608859 Free PMC article.
-
Role of previous infection with SARS-CoV-2 in protecting against omicron reinfections and severe complications of COVID-19 compared to pre-omicron variants: a systematic review.BMC Infect Dis. 2023 Jun 26;23(1):432. doi: 10.1186/s12879-023-08328-3. BMC Infect Dis. 2023. PMID: 37365490 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
