This is a preprint.
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid utilization regulator
- PMID: 37662244
- PMCID: PMC10473576
- DOI: 10.1101/2023.08.22.554309
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid utilization regulator
Update in
-
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid metabolism regulator.PLoS Genet. 2024 Jan 24;20(1):e1011143. doi: 10.1371/journal.pgen.1011143. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38266039 Free PMC article.
Abstract
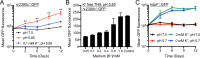
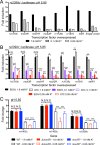
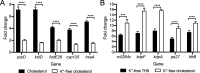
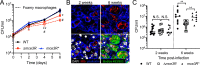
How bacterial response to environmental cues and nutritional sources may be integrated in enabling host colonization is poorly understood. Exploiting a reporter-based screen, we discovered that overexpression of Mycobacterium tuberculosis (Mtb) lipid utilization regulators altered Mtb acidic pH response dampening by low environmental potassium (K+). Transcriptional analyses unveiled amplification of Mtb response to acidic pH in the presence of cholesterol, a major carbon source for Mtb during infection, and vice versa. Strikingly, deletion of the putative lipid regulator mce3R resulted in loss of augmentation of (i) cholesterol response at acidic pH, and (ii) low [K+] response by cholesterol, with minimal effect on Mtb response to each signal individually. Finally, the ∆mce3R mutant was attenuated for colonization in a murine model that recapitulates lesions with lipid-rich foamy macrophages. These findings reveal critical coordination between bacterial response to environmental and nutritional cues, and establish Mce3R as a crucial integrator of this process.
Keywords: C3HeB/FeJ; Mycobacterium tuberculosis; acidic pH; cholesterol; environmental cues; foamy macrophages; lipids; mce3R; potassium.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures



Similar articles
-
Mycobacterium tuberculosis response to cholesterol is integrated with environmental pH and potassium levels via a lipid metabolism regulator.PLoS Genet. 2024 Jan 24;20(1):e1011143. doi: 10.1371/journal.pgen.1011143. eCollection 2024 Jan. PLoS Genet. 2024. PMID: 38266039 Free PMC article.
-
Cholesterol metabolism and intrabacterial potassium homeostasis are intrinsically related in Mycobacterium tuberculosis.bioRxiv [Preprint]. 2024 Nov 11:2024.11.10.622811. doi: 10.1101/2024.11.10.622811. bioRxiv. 2024. Update in: PLoS Pathog. 2025 May 22;21(5):e1013207. doi: 10.1371/journal.ppat.1013207. PMID: 39605342 Free PMC article. Updated. Preprint.
-
Targeting Mycobacterium tuberculosis response to environmental cues for the development of effective antitubercular drugs.PLoS Biol. 2021 Jul 28;19(7):e3001355. doi: 10.1371/journal.pbio.3001355. eCollection 2021 Jul. PLoS Biol. 2021. PMID: 34319985 Free PMC article.
-
Mycobacterium tuberculosis Infection-Driven Foamy Macrophages and Their Implications in Tuberculosis Control as Targets for Host-Directed Therapy.Front Immunol. 2020 May 12;11:910. doi: 10.3389/fimmu.2020.00910. eCollection 2020. Front Immunol. 2020. PMID: 32477367 Free PMC article. Review.
-
Not too fat to fight: The emerging role of macrophage fatty acid metabolism in immunity to Mycobacterium tuberculosis.Immunol Rev. 2021 May;301(1):84-97. doi: 10.1111/imr.12952. Epub 2021 Feb 8. Immunol Rev. 2021. PMID: 33559209 Review.
References
-
- World Health Organization. (2022). Global tuberculosis report. https://www.who.int/publications/i/item/9789240061729. (Last accessed 18 August 2023.)
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
