This is a preprint.
Neuroimaging-AI endophenotypes reveal underlying mechanisms and genetic factors contributing to progression and development of four brain disorders
- PMID: 37662256
- PMCID: PMC10473785
- DOI: 10.1101/2023.08.16.23294179
Neuroimaging-AI endophenotypes reveal underlying mechanisms and genetic factors contributing to progression and development of four brain disorders
Update in
-
Neuroimaging endophenotypes reveal underlying mechanisms and genetic factors contributing to progression and development of four brain disorders.Nat Biomed Eng. 2025 Nov;9(11):1920-1937. doi: 10.1038/s41551-025-01412-w. Epub 2025 Jun 6. Nat Biomed Eng. 2025. PMID: 40481237
Abstract
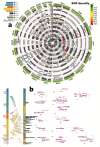
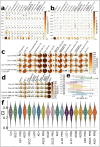
Recent work leveraging artificial intelligence has offered promise to dissect disease heterogeneity by identifying complex intermediate brain phenotypes, called dimensional neuroimaging endophenotypes (DNEs). We advance the argument that these DNEs capture the degree of expression of respective neuroanatomical patterns measured, offering a dimensional neuroanatomical representation for studying disease heterogeneity and similarities of neurologic and neuropsychiatric diseases. We investigate the presence of nine DNEs derived from independent yet harmonized studies on Alzheimer's disease, autism spectrum disorder, late-life depression, and schizophrenia in the UK Biobank study. Phenome-wide associations align with genome-wide associations, revealing 31 genomic loci (P-value<5×10-8/9) associated with the nine DNEs.The nine DNEs, along with their polygenic risk scores, significantly enhanced the predictive accuracy for 14 systemic disease categories, particularly for conditions related to mental health and the central nervous system, as well as mortality outcomes. These findings underscore the potential of the nine DNEs to capture the expression of disease-related brain phenotypes in individuals of the general population and to relate such measures with genetics, lifestyle factors, and chronic diseases.
Conflict of interest statement
Competing Interests Statement None
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
